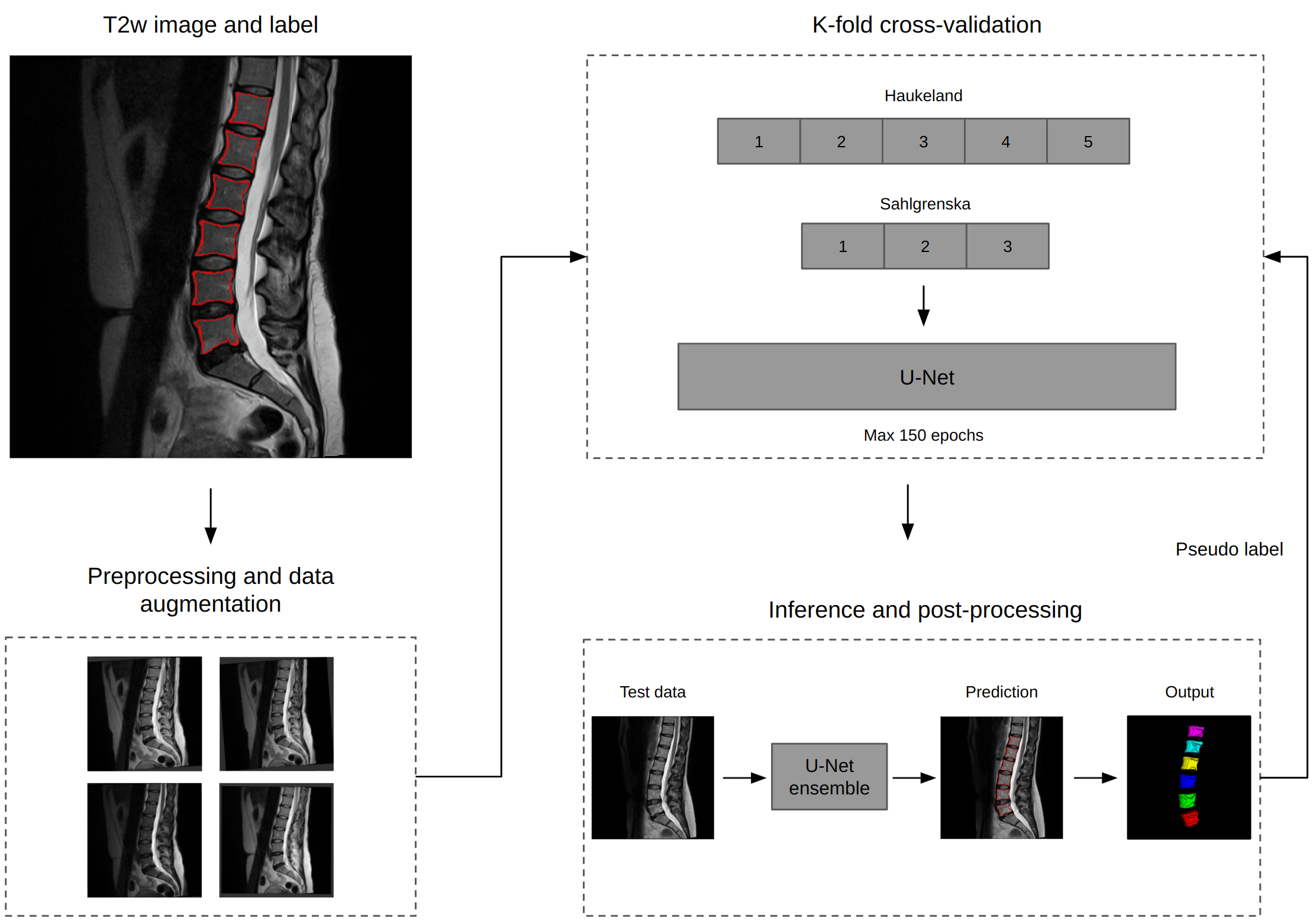
Multi-Center CNN-based spine segmentation from T2w MRI using small amounts of data
Pre-trained weights and exported models for our spine segmentation project. The source code, designed to reproduce our test results and facilitate training and running inference on your own data, is available on GitHub: https://github.com/MMIV-ML/fastMONAI/tree/master/research
Requirements
Last checked and validated with fastMONAI version 0.3.2. Please ensure that you have the correct version of fastMONAI installed to guarantee the correct operation of the model.
Data
Haukeland University Hospital (HUH): 15 T2-weighted MRI scans
Scanner Specifications
- Type: Siemens MAGNETOM Avanto 1.5 T
- Slice Thickness: 4 mm
- Field of View (FOV): 300x300 mm^2
- Matrix: 384x269
- Interslice Gap: 0.4 mm
- Repetition Time: 3700 ms
- Echo Time: 87 ms
- Number of Acquisitions: 2
Sahlgren-ska University Hospital (SUH): 10 T2-weighted MRI scans
Scanner Specifications
- Type: Siemens MAGNETOM Aera 1.5 T
- Slice Thickness: 3.5 mm
- Field of View (FOV): 300x300 mm^2
- Matrix: 384x384
- Interslice Gap: 0.7 mm
- Repetition Time: 3500 ms
- Echo Time: 95 ms
- Number of Acquisitions: 1
Data Annotation
- Process: Manual segmentation of vertebrae
- Experts: Conducted by an MRI expert with consensus from a senior radiologist
- Software: ITK-SNAP
- Scope: Segmented vertebrae in all slices, including bone marrow but excluding vertebral cortex
External Data Evaluation
- Source: Annotated T2-weighted MR images of the Lower Spine
- Scan Specifications: 23 scans, 1.5 T in sagittal orientation
- Segments: Seven vertebrae are manually segmented on each image (Th11–L5)
- Modification: Th11 manually removed for consistency with the annotation for SUH and HUH.
Inference API (serverless) does not yet support fastMONAI models for this pipeline type.