path
stringlengths 8
399
| content_id
stringlengths 40
40
| detected_licenses
sequence | license_type
stringclasses 2
values | repo_name
stringlengths 6
109
| repo_url
stringlengths 25
128
| star_events_count
int64 0
52.9k
| fork_events_count
int64 0
7.07k
| gha_license_id
stringclasses 9
values | gha_event_created_at
timestamp[us] | gha_updated_at
timestamp[us] | gha_language
stringclasses 28
values | language
stringclasses 1
value | is_generated
bool 1
class | is_vendor
bool 1
class | conversion_extension
stringclasses 17
values | size
int64 317
10.5M
| script
stringlengths 245
9.7M
| script_size
int64 245
9.7M
|
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
/Python/py/pyds_subfolders/ipynb/img2data.ipynb | a0759dec6d7981e65ce9b83a27454698ed2238c8 | [] | no_license | astrodoo/CodeExr | https://github.com/astrodoo/CodeExr | 0 | 0 | null | null | null | null | Jupyter Notebook | false | false | .py | 96,563 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Python 2
# language: python
# name: python2
# ---
# # Example of using img2data routine for collecting data position from image plot
#
# ### NOTE that magic function should be set to the one that allows interactive plotting such as '%matplotlib notebook'
#
# * First, you should click four boundary points.
# --> click first two points for setting minimum x & maximum x and next two points for setting minimum y & maximum y.
# * Second, now you can click data points repeatedly.
# * Third, when you fininshed clicking all data point, then you should finalize it with clicking 'Stop Interaction' Mark.
# * All data points will be saved in self.xdata & self.ydata in the type of 'list'.
# ## 1) Linear scale plot
# +
""" This is not interactive mode. """
import matplotlib.image as mpimg
import matplotlib.pyplot as plt
import os
# %matplotlib inline
pyfig = os.environ['PYTHONDATA']
img = mpimg.imread(pyfig+'testlinear.png')
fig, ax = plt.subplots(figsize=(8,8.*img.shape[0]/img.shape[1])) ## note that imgsz[0] represents y-axis size
imgplot = ax.imshow(img)
# +
""" Now, it is interactive mode and you can collect all data point by clicking mouse.
Sometimes, transitioning from '%matplotlib inline' to '%matplotlib notebook' seems not work smoothly.
In this case, please just run this one more time, then it would work.
"""
from pyds.img2data import readimg
import matplotlib.pyplot as plt
import matplotlib.image as mpimg
import os
# !!!!! important !!!!!
# %matplotlib notebook
pyfig = os.environ['PYTHONDATA']
img = mpimg.imread(pyfig+'testlinear.png')
fig, ax = plt.subplots(figsize=(8,8.*img.shape[0]/img.shape[1]))
imgplot = ax.imshow(img)
# x/y range in the plot accorindg to the position you set for boundary
xlim = [2.,6.]
ylim = [4.5,6.]
data = readimg(imgplot,ax,xlim=xlim,ylim=ylim)
# +
""" no lines
"""
from pyds.img2data import readimg
import matplotlib.pyplot as plt
import matplotlib.image as mpimg
import os
# !!!!! important !!!!!
# %matplotlib notebook
pyfig = os.environ['PYTHONDATA']
img = mpimg.imread(pyfig+'testlinear.png')
fig, ax = plt.subplots(figsize=(8,8.*img.shape[0]/img.shape[1]))
imgplot = ax.imshow(img)
# x/y range in the plot accorindg to the position you set for boundary
xlim = [2.,6.]
ylim = [4.5,6.]
data = readimg(imgplot,ax,xlim=xlim,ylim=ylim, noline=True)
# +
""" printing the data points you collected """
print data.xdata,data.ydata
# +
""" check the data points with plot """
import matplotlib.pyplot as plt
# %matplotlib inline
fig,ax = plt.subplots(figsize=(8,8))
ax.plot(data.xdata,data.ydata,'o')
ax.set_xlim([0.6,8.5])
ax.set_ylim([4.,6.3])
# -
# ## 2) Log scale plot
# +
""" This is not interactive mode. """
import matplotlib.image as mpimg
import matplotlib.pyplot as plt
import os
# %matplotlib inline
pyfig = os.environ['PYTHONDATA']
img = mpimg.imread(pyfig+'testlog.png')
fig, ax = plt.subplots(figsize=(8,8.*img.shape[0]/img.shape[1]))
imgplot = ax.imshow(img)
# +
""" Now, it is interactive mode and you can collect all data point by clicking mouse.
Sometimes, transitioning from '%matplotlib inline' to '%matplotlib notebook' seems not work smoothly.
In this case, please just run this one more time, then it would work.
"""
from pyds.img2data import readimg
import matplotlib.pyplot as plt
import matplotlib.image as mpimg
import os
# !!!!! important !!!!!
# %matplotlib notebook
pyfig = os.environ['PYTHONDATA']
img = mpimg.imread(pyfig+'testlog.png')
fig, ax = plt.subplots(figsize=(8,8.*img.shape[0]/img.shape[1]))
imgplot = ax.imshow(img)
# x/y range in the plot accorindg to the position you set for boundary
xlim = [1e-6,1.]
ylim = [1e-6,1.]
data = readimg(imgplot,ax,xlim=xlim,ylim=ylim,xlog=True,ylog=True)
# +
""" printing the data points you collected """
print data.xdata,data.ydata
# +
""" check the data points with plot """
import matplotlib.pyplot as plt
# %matplotlib inline
fig,ax = plt.subplots(figsize=(8,8))
ax.loglog(data.xdata,data.ydata,'o')
ax.set_xlim([1e-6,1])
ax.set_ylim([1e-6,9.])
| 4,294 |
/winter21/2_estimate_real_user_pp/2_error_calc.ipynb | d7c3f9590e2bd43ce7f1dd7ae6a7c6ec6a265cee | [
"MIT"
] | permissive | the-data-science-union/dsu-mlpp | https://github.com/the-data-science-union/dsu-mlpp | 0 | 2 | MIT | 2021-03-04T03:49:42 | 2021-03-04T01:27:13 | Jupyter Notebook | Jupyter Notebook | false | false | .py | 15,174 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# + cell_id="00000-47a1f3f7-7379-4e2b-b5f6-1868e003b07d" deepnote_cell_type="code"
import sys
sys.path.append('../..')
import pandas as pd
from pymongo import UpdateOne
from pymongo import MongoClient
from tqdm import tqdm
import numpy as np
from exploration.config import mongo_inst
from mlpp.data_collection.sample import osuDumpSampler
import datetime
from datetime import datetime
import pprint
import matplotlib.pyplot as plt
import pickle
from fastdtw import fastdtw
from scipy.spatial.distance import euclidean
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
# + [markdown] cell_id="00001-a0180432-996a-44e2-8aa7-aad9ffb37747" deepnote_cell_type="markdown"
# # Data Retrievel
# + [markdown] cell_id="00002-6af4c1e8-e780-454b-807a-6e5d1ca4a4f7" deepnote_cell_type="markdown"
# ### Find users that have more than 50 data points & creates a new list of user ids
# + cell_id="00003-a1d32fed-1720-43a1-a24b-bc2c04c075f7" deepnote_cell_type="code"
client = MongoClient(port=27017)
top_db = mongo_inst["osu_top_db"]
user_ids = list(map(lambda c: c['_id'], top_db['osu_user_stats'].find({}, {})))
# + cell_id="00004-e662fc18-9e69-46d7-b71d-97d378989da0" deepnote_cell_type="code"
big_user_ids = []
for i in user_ids:
datapts = len(list(top_db.osu_track_updates.find({"user_id": i}, {"date":1})))
if datapts > 50: #arbitrary number
big_user_ids.append(i)
# + [markdown] cell_id="00005-82400054-4033-4b05-b36c-3425f2ff66a4" deepnote_cell_type="markdown"
#
# ### Find the date & real_pp and then sort by date
#
# + cell_id="00006-023f7d5d-3588-456d-8920-9fddcfd2798d" deepnote_cell_type="code"
real_pp = {}
for i in big_user_ids:
real_pp[i] = list(top_db.osu_track_updates.find( {"user_id": i}, {"_id":0, "timestamp": 1, "pp_raw":1}))
real_pp[i].sort(key = lambda x:x["timestamp"])
# + [markdown] cell_id="00007-b2e2b0f9-c95a-4808-8a5e-415abbe9e1ca" deepnote_cell_type="markdown"
# ### Find the date and est_user_pp and then sort by date
# + cell_id="00008-b87d7625-b4e0-4562-80c3-164b58c8f6c5" deepnote_cell_type="code"
est_pp = {}
for i in big_user_ids:
est_pp[i] = list(top_db.osu_scores_high.find( {"user_id": i}, {"_id":0, "date": 1, "mlpp.est_user_pp":1}))
est_pp[i].sort(key = lambda x:x["date"])
# + [markdown] cell_id="00009-1bec2fa3-4b58-4678-89a7-f524bf439579" deepnote_cell_type="markdown"
# ### Find the overlap time periods
# + cell_id="00010-e44690a7-1e17-4199-9b91-017c9eae8d0d" deepnote_cell_type="code"
time_comparison = {}
for i in big_user_ids:
earliest_est_date = est_pp[i][0]["date"]
earliest_real_date = real_pp[i][0]["timestamp"]
latest_est_date = est_pp[i][-1]["date"]
latest_real_date = real_pp[i][-1]["timestamp"]
if earliest_est_date >= earliest_real_date:
start = earliest_est_date
else:
start = earliest_real_date
if latest_est_date <= latest_real_date:
end = latest_est_date
else:
end = latest_real_date
time_comparison[i] = {"start": start, "end": end}
# + [markdown] cell_id="00011-55096b7d-92a7-4755-b0a2-c6cec20df3c5" deepnote_cell_type="markdown"
# ### Clean the data for users who do not have time overlap between real and est
# + cell_id="00012-45bc490f-4227-426b-aa56-b5705cbc8a3c" deepnote_cell_type="code"
bad_users = []
for user in time_comparison:
if time_comparison[user]["start"] >= time_comparison[user]["end"]:
bad_users.append(user)
big_user_ids = [ele for ele in big_user_ids if ele not in bad_users]
for user in bad_users:
del real_pp[user]
del est_pp[user]
del time_comparison[user]
# + [markdown] cell_id="00013-14501696-82ad-4bfe-b3c4-89f5b5ef8746" deepnote_cell_type="markdown"
# ### Create tables to store the data
# + cell_id="00014-ed01aed9-8e81-4a2c-a0f2-34a5eb71ba7e" deepnote_cell_type="code"
def CREATE_REAL_USER_TABLE (one_user_id):
real_start_time = []
real_pp_points = []
for i in real_pp[one_user_id]:
real_start_time.append(i["timestamp"])
real_pp_points.append(i["pp_raw"])
real_table = np.column_stack((real_start_time, real_pp_points))
return real_table
# + cell_id="00015-53bc8cf3-9524-43fb-a9d9-b5e5a2d0fa1e" deepnote_cell_type="code"
real_table_for_all = {}
for user in big_user_ids:
real_table_for_all[user] = CREATE_REAL_USER_TABLE(user)
# + cell_id="00016-678ede0b-3301-4ec5-85ba-22b516ffbc86" deepnote_cell_type="code"
def CREATE_EST_USER_TABLE (one_user_id):
est_start_time = []
est_pp_points = []
for i in est_pp[one_user_id]:
est_start_time.append(i["date"])
est_pp_points.append(i["mlpp"]["est_user_pp"])
est_table = np.column_stack((est_start_time, est_pp_points))
return est_table
# + cell_id="00017-9abf705d-60cd-45a8-8468-edd8e4e4df88" deepnote_cell_type="code"
est_table_for_all = {}
for user in big_user_ids:
est_table_for_all[user] = CREATE_EST_USER_TABLE(user)
# + [markdown] cell_id="00018-2bf0862f-e5a2-4fea-81c5-127b440d4b53" deepnote_cell_type="markdown"
# # Calculate error
# + cell_id="00019-bcd723ef-b264-4c24-8ad1-a54c2e6a43ba" deepnote_cell_type="code"
def GET_REAL_NPOINTS(n, user):
real_within = real_table_for_all[user][real_table_for_all[user][:,0] >= time_comparison[user]["start"]]
real_within = real_within[real_within[:,0] <= time_comparison[user]["end"]]
real_xp = real_within[:,0]
real_fp = real_within[:,1]
begin = real_xp[0]
end = real_xp[-1]
real_date_list = []
delta = (end - begin)/n
for i in range(1, n + 1):
real_date_list.append((begin+i*delta).timestamp())
k = 0
for i in real_xp:
real_xp[k] = i.timestamp()
k+=1
real_npoints = np.interp(real_date_list,list(real_xp),list(real_fp))
return real_npoints
# + cell_id="00020-cf1e6da1-95b6-4f82-a660-bfefbec89036" deepnote_cell_type="code"
def GET_EST_NPOINTS(n, user):
est_within = est_table_for_all[user][est_table_for_all[user][:,0] >= time_comparison[user]["start"]]
est_within = est_within[est_within[:,0] <= time_comparison[user]["end"]]
xp = est_within[:,0]
fp = est_within[:,1]
begin = xp[0]
end = xp[-1]
date_list = []
delta = (end - begin)/n
for i in range(1, n + 1):
date_list.append((begin+i*delta).timestamp())
k = 0
for i in xp:
xp[k] = i.timestamp()
k+=1
est_npoints = np.interp(date_list,list(xp),list(fp))
return est_npoints
# + [markdown] cell_id="00021-6eb6fb56-4f4a-4648-8cfa-a8e6b39f4def" deepnote_cell_type="markdown"
# ### MSE
# + cell_id="00022-5948a738-e68a-41e2-a725-461c9b9d4d57" deepnote_cell_type="code"
def GET_MSE(n, user):
real_points = GET_REAL_NPOINTS(n, user)
est_points = GET_EST_NPOINTS(n, user)
mse_for_one = (np.square(real_points - est_points)).mean()
return mse_for_one
# + cell_id="00023-3d3186ac-198d-476c-9190-3119458623d7" deepnote_cell_type="code"
mse_for_all = {}
for user in big_user_ids:
mse_for_all[user] = GET_MSE(50, user)
# + [markdown] cell_id="00024-6457df83-fd38-4d7b-9f8c-81f22ffafdda" deepnote_cell_type="markdown"
# ### Difference in area
# + cell_id="00025-ca012959-4ceb-4bf6-b3c9-5053e83d6803" deepnote_cell_type="code"
def GET_AREA(n, user, intervals):
#interval is an arbitrary number, it is the width of the small rectangles, we used 1000 before
a = time_comparison[user]["start"].timestamp()
b = time_comparison[user]["end"].timestamp()
dx = (b - a) / intervals
x_midpoint = np.linspace(dx / 2, b - dx / 2, intervals)
total_area_between_curves = 0
real_nPoints = GET_REAL_NPOINTS(n, user)
est_nPoints = GET_EST_NPOINTS(n, user)
for i in range(0, n):
real_midpoint_riemann = real_nPoints[i] * dx
est_midpoint_riemann = est_nPoints[i] * dx
area_between_curves_one_point = abs(real_midpoint_riemann - est_midpoint_riemann)
total_area_between_curves += area_between_curves_one_point
total_area_between_curves = total_area_between_curves / 86400
return total_area_between_curves
# + cell_id="00026-206935a4-9084-49eb-98c5-d8b5aed019b4" deepnote_cell_type="code"
area_for_all = {}
for user in big_user_ids:
area_for_all[user] = GET_AREA(10000, user, 1000)
# + [markdown] cell_id="00027-871efcd9-fd15-49f0-b128-58e6aedeca38" deepnote_cell_type="markdown"
# ### Dynamic Time Warping
# + cell_id="00028-bc49b4de-0208-4200-85bb-6405b782993e" deepnote_cell_type="code"
def GET_DTW(user):
real_within = real_table_for_all[user][real_table_for_all[user][:,0] >= time_comparison[user]["start"]]
real_within = real_within[real_within[:,0] <= time_comparison[user]["end"]]
est_within = est_table_for_all[user][est_table_for_all[user][:,0] >= time_comparison[user]["start"]]
est_within = est_within[est_within[:,0] <= time_comparison[user]["end"]]
distance = fastdtw(real_within[:,1], est_within[:,1], dist=euclidean)[0]
return distance
# + cell_id="00029-a5fbb45b-19d3-4867-8c2b-3c7c6e773146" deepnote_cell_type="code"
dtw_for_all = {}
for user in big_user_ids:
dtw_for_all[user] = GET_DTW(user)
# + [markdown] cell_id="00030-734b0edf-f501-4a1b-8160-d78e27a0910e" deepnote_cell_type="markdown"
# ### Merge errors calculated through three methods to one dataframe
# + cell_id="00031-cda7cfbe-bf3c-4932-bd22-90112d0c9669" deepnote_cell_type="code"
error_df = pd.DataFrame({'mse_for_all':pd.Series(mse_for_all),'area_for_all':pd.Series(area_for_all), 'dtw_for_all':pd.Series(dtw_for_all)})
error_df
ion3*Recall3/(Precision3+Recall3)
F1_3
params3
# **通过对比,不难发现:3.3的结果要略微好于3.2的结果,因此我们选择3.3的模型:**
# `Intercept 45.272344`
# `Petal_Length -5.754532`
# `Petal_Width -10.446700`
# `dtype: float64`
# ### 3.7 拟合
#
# **按result3(3.3)结果拟合**
beta0 = -params3['Intercept']/params3['Petal_Width']
beta1 = -params3['Petal_Length']/params3['Petal_Width']
df_subset.describe()
df_new = pd.DataFrame({"Petal_Length": np.random.randn(20)*0.5 +5,
"Petal_Width" :np.random.randn(20)*0.5+1.7})
df_new["P-Species"] = result3.predict(df_new)
df_new.head()
df_new["Species"] = (df_new["P-Species"] > 0.5).astype(int)
df_new.head()
# +
fig, ax = plt.subplots(1,1,figsize=(8,4))
ax.plot(df_subset[df_subset.Species == 0].Petal_Length.values,
df_subset[df_subset.Species == 0].Petal_Width.values, 's', label='virginica')
ax.plot(df_new[df_new.Species == 0].Petal_Length.values,
df_new[df_new.Species == 0].Petal_Width.values,
'o', markersize=10, color="steelblue", label='virginica (pred.)')
ax.plot(df_subset[df_subset.Species == 1].Petal_Length.values,
df_subset[df_subset.Species == 1].Petal_Width.values, 's', label='versicolor')
ax.plot(df_new[df_new.Species == 1].Petal_Length.values,
df_new[df_new.Species == 1].Petal_Width.values,
'o', markersize=10, color="green", label='versicolor (pred.)')
_x = np.array([4.0,6.1])
ax.plot(_x, beta0 + beta1 * _x, 'k')
ax.set_xlabel('Petal length')
ax.set_ylabel('Petal width')
ax.legend(loc=2)
fig.tight_layout()
# -
# ## 4.Time series
# !head -n 5 temperature_outdoor_2014.tsv
# !wc -l temperature_outdoor_2014.tsv
df = pd.read_csv("temperature_outdoor_2014.tsv", header=None, delimiter="\t",names=["time","temp"])
df.time = pd.to_datetime(df.time, unit="s")
df.head()
df = df.set_index("time").resample("H").mean()
df.head()
df.head()
df.tail()
df_march = df[df.index.month == 3]
df_april = df[df.index.month == 4]
df_march.plot(figsize=(12,4));
plt.scatter(df_april[1:], df_april[:-1]);
df_april.tail()
df_april[:-1].tail()
plt.scatter(df_april[:-1], df_april[1:]);
plt.scatter(df_april[2:], df_april[:-2]);
plt.scatter(df_april[24:], df_april[:-24]);
plt.scatter(df_april[:-24], df_april[24:]);
# +
# 差分,可使非平稳时间序列变为平稳的;可以消除序列相关(自相关)。
fig, axes = plt.subplots(1,4,figsize=(15,4))
smg.tsa.plot_acf(df_march.temp, lags=72, ax=axes[0])
smg.tsa.plot_acf(df_march.temp.diff().dropna(),lags=72,ax=axes[1])
smg.tsa.plot_acf(df_march.temp.diff().diff().dropna(), lags=72, ax=axes[2])
smg.tsa.plot_acf(df_march.temp.diff().diff().diff().dropna(), lags=72, ax=axes[3])
fig.tight_layout()
# plot_acf在时间序列上可视化自相关;pacf偏相关。
# +
model = sm.tsa.AR(df_march.temp)
result = model.fit(72)
# 72小时
# AR模型(自回归模型)
# +
sm.stats.durbin_watson(result.resid)
# DW=2,非自相关
# +
fig, ax = plt.subplots(1,1,figsize=(8,3))
smg.tsa.plot_acf(result.resid, lags=72, ax=ax)
fig.tight_layout()
# +
fig, ax = plt.subplots(1,1,figsize=(12,4))
ax.plot(df_march.index.values[-72:], df_march.temp.values[-72:],label="train data")
ax.plot(df_april.index.values[:72], df_april.temp.values[:72], label="actual outcome")
ax.plot(pd.date_range("2014-04-01", "2014-04-04", freq="H").values,
result.predict("2014-04-01", "2014-04-04"), label="predicted outcome")
ax.legend()
fig.tight_layout()
# -
# ## 5.补充-ARIMA模型
# ### 5.1 获取数据
dta=[10930,10318,10595,10972,7706,6756,9092,10551,9722,10913,11151,8186,6422,
6337,11649,11652,10310,12043,7937,6476,9662,9570,9981,9331,9449,6773,6304,9355,
10477,10148,10395,11261,8713,7299,10424,10795,11069,11602,11427,9095,7707,10767,
12136,12812,12006,12528,10329,7818,11719,11683,12603,11495,13670,11337,10232,
13261,13230,15535,16837,19598,14823,11622,19391,18177,19994,14723,15694,13248,
9543,12872,13101,15053,12619,13749,10228,9725,14729,12518,14564,15085,14722,
11999,9390,13481,14795,15845,15271,14686,11054,10395]
dta=pd.Series(dta)
dta.index = pd.Index(sm.tsa.datetools.dates_from_range('2001','2090'))
dta.plot(figsize=(12,8))
# 非平稳序列,差分平稳后才能继续分析。
# ### 5.2 差分
fig = plt.figure(figsize=(12,8))
ax1= fig.add_subplot(111)
diff1 = dta.diff(1)
diff1.plot(ax=ax1);
fig = plt.figure(figsize=(12,8))
ax1= fig.add_subplot(111)
#diff2 = dta.diff().diff()
diff2 = dta.diff(2)
diff2.plot(ax=ax1);
# 一阶差分和二阶差分结果相差不大,所以一阶差分即可。$d=1$
# ### 5.3 确定$pq$
dta= dta.diff(1).dropna()
fig = plt.figure(figsize=(12,8))
ax1=fig.add_subplot(211)
fig = smg.tsa.plot_acf(dta,lags=40,ax=ax1)
ax2 = fig.add_subplot(212)
fig = smg.tsa.plot_pacf(dta,lags=40,ax=ax2)
# ?sm.tsa.ARMA().fit()
# +
arma_mod20 = sm.tsa.ARMA(dta,(7,0)).fit([7,0])
print(arma_mod20.aic,arma_mod20.bic,arma_mod20.hqic);
arma_mod30 = sm.tsa.ARMA(dta,(0,1)).fit()
print(arma_mod30.aic,arma_mod30.bic,arma_mod30.hqic);
arma_mod40 = sm.tsa.ARMA(dta,(7,1)).fit()
print(arma_mod40.aic,arma_mod40.bic,arma_mod40.hqic); # 最佳
arma_mod40 = sm.tsa.ARMA(dta,(7,3)).fit()
print(arma_mod40.aic,arma_mod40.bic,arma_mod40.hqic);
arma_mod50 = sm.tsa.ARMA(dta,(8,0)).fit([8,0])
print(arma_mod50.aic,arma_mod50.bic,arma_mod50.hqic);
# -
# ### 5.4 检验
# #### 5.4.1 残差自相关图
# +
fig = plt.figure(figsize=(12,8))
ax1 = fig.add_subplot(211)
fig = smg.tsa.plot_acf(arma_mod40.resid.values.squeeze(), lags=40, ax=ax1)
#fig = smg.tsa.plot_acf(arma_mod40.resid, lags=40, ax=ax1)
# >>> x = np.array([[[0], [1], [2]]])
# >>> x.shape
# (1, 3, 1)
# >>> np.squeeze(x).shape
# (3,)
# >>> np.squeeze(x, axis=(2,)).shape
# (1, 3)
ax2 = fig.add_subplot(212)
fig = smg.tsa.plot_pacf(arma_mod40.resid, lags=40, ax=ax2)
# -
# #### 5.4.3 DW检验
# +
sm.stats.durbin_watson(arma_mod40.resid)
# 不存在自相关
# -
# #### 5.4.4 正态检验
fig = plt.figure(figsize=(12,8))
ax = fig.add_subplot(111)
fig = smg.qqplot(arma_mod40.resid, line='q', ax=ax, fit=True)
# #### 5.4.5 LBQ检验
r,q,p = sm.tsa.acf(arma_mod40.resid.values.squeeze(), qstat=True)
data = np.c_[range(1,41), r[1:], q, p]
table = pd.DataFrame(data, columns=['lag', "AC", "Q", "Prob(>Q)"])
print(table.set_index('lag'))
# ### 5.5 预测
predict_sunspots = arma_mod40.predict('2090', '2100', dynamic=True)
print(predict_sunspots)
fig, ax = plt.subplots(figsize=(12, 8))
ax = dta.ix['2001':].plot(ax=ax)
predict_sunspots.plot(ax=ax)
| 15,936 |
/Stack/Stack_Next_Greater_Frequency_Element.ipynb | 915e56df86279fda976d91193e8075e5f4ba4512 | [] | no_license | bilalsp/DataStructures | https://github.com/bilalsp/DataStructures | 1 | 0 | null | null | null | null | Jupyter Notebook | false | false | .py | 3,308 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# +
import scipy.integrate as integrate
import matplotlib.pyplot as plt
import numpy as np
from scipy import interpolate
from math import *
# +
N_grid=1000
L=10
dw=0.1
# -
t=np.linspace(0, L, N_grid)
h=t[2]-t[1]
# +
C0=np.array([0.5]*N_grid)
C0[300:500]=0.9
phi_0=np.arccos(C0)
k0=np.divide(np.array([2*dw]*N_grid), np.sin(phi_0))
plt.figure(figsize=(5, 5))
plt.plot(t, k0)
plt.show()
# -
dphi_0=np.diff(phi_0)/h
dphi_0=np.append(dphi_0[1], dphi_0)
plt.figure(figsize=(5,5))
plt.plot(t, dphi_0, 'b')
plt.plot(t, [2*dw]*N_grid, 'r--')
plt.plot(t, [-2*dw]*N_grid, 'r--')
plt.show()
# +
k=np.divide(np.array([2*dw]*N_grid)-dphi_0, np.sin(phi_0))
k_f=interpolate.interp1d(t, k, bounds_error=False, fill_value="extrapolate")
plt.figure(figsize=(5, 5))
plt.plot(t, k)
plt.plot(t, k_f(t))
plt.show()
# -
def deriv_z(z, t2, dw, k_f):
return 2*dw-sin(z)*k_f(t2)
# +
theta = integrate.odeint(deriv_z, phi_0[1], t, (dw, k_f,))
theta=theta.ravel()
plt.plot(t, phi_0)
plt.plot(t, theta)
plt.show()
# +
C0_hat=np.cos(theta)
plt.figure(figsize=(10,10))
plt.grid(True)
plt.plot(t, C0, 'r')
plt.plot(t, C0_hat, 'b')
plt.show()
# +
dphi_hat=np.diff(theta)/h
dphi_hat=np.append(dphi_hat[1], dphi_hat)
k_hat=np.divide(np.array([2*dw]*N_grid)-dphi_hat, np.sin(theta))
for i in range(N_grid):
if (abs(k_hat[i])>1):
k_hat[i]=nan
plt.figure(figsize=(10,10))
plt.grid(True)
plt.plot(t, k0, 'r')
plt.plot(t, k_hat, 'b')
plt.show()
# -
y = pca_values[:,1:2]
#z = pca_values[:2:3]
plt.scatter(x,y)
finalDf = pd.concat([pd.DataFrame(pca_values[:,0:2],columns=['pc1','pc2']), wine[['Type']]], axis = 1)
import seaborn as sns
sns.scatterplot(data=finalDf,x='pc1',y='pc2',hue='Type')
# # Heirarchial Clustering
# Normalization function
def norm_func(i):
x = (i-i.min())/(i.max()-i.min())
return (x)
# Normalized data frame (considering the numerical part of data)
df_norm = norm_func(wine.iloc[:,1:])
# create dendrogram
dendrogram = sch.dendrogram(sch.linkage(df_norm, method='single'))
# create clusters
hc = AgglomerativeClustering(n_clusters=4, affinity = 'euclidean', linkage = 'single')
# save clusters for chart
y_hc = hc.fit_predict(df_norm)
Clusters=pd.DataFrame(y_hc,columns=['Clusters'])
df_norm['h_clusterid'] = hc.labels_
df_norm['h_clusterid'].head()
pd.concat([wine,df_norm['h_clusterid']],axis=1)
# # K-Means
# Normalization function
from sklearn.preprocessing import StandardScaler
scaler = StandardScaler()
scaled_wine_df = scaler.fit_transform(wine.iloc[:,1:])
ks = range(1, 10)
inertias = []
for k in ks:
model = KMeans(n_clusters=k)
model.fit(pca_values[:,:3])
inertias.append(model.inertia_)
plt.plot(ks, inertias, '-o', color='black')
plt.xlabel('number of clusters, k')
plt.ylabel('inertia')
plt.xticks(ks)
plt.show()
# # The above Scree plot shows that after k=3 there is slow decrease in the inertia.Hence,the number of clusters is 3 this is same as the number of clusters obtained from the original data.¶
scaler = StandardScaler()
scaled_wine_df = scaler.fit_transform(pca_values[:,:3])
#Build Cluster algorithm
from sklearn.cluster import KMeans
clusters_new = KMeans(4, random_state=42)
clusters_new.fit(scaled_wine_df)
clusters_new.labels_
#Assign clusters to the data set
wine['clusterid_new'] = clusters_new.labels_
#these are standardized values.
clusters_new.cluster_centers_
wine.groupby('clusterid_new').agg(['mean']).reset_index()
wine
):
#print(event)
if event.type==pygame.QUIT:
pygame.quit()
quit()
gameDisplay.fill(white)
largeText = pygame.font.Font('freesansbold.ttf',115)
TextSurf, TextRect = text_objects("Car Dash", largeText)
TextRect.center = ((display_width/2),(display_height/2))
gameDisplay.blit(TextSurf, TextRect)
button("GO!",150,450,100,50,green,bgreen, "PLAY")
button("QUIT!",550,450,100,50,red, bred, "QUIT")
pygame.display.update()
clock.tick(15)
def game_loop():
global pause
x = (display_width * 0.45)
y = (display_height * 0.8)
x_change = 0
thing_startx=random.randrange(0, display_width)
thing_starty=-600
thing_speed = 5
thing_width=100
thing_height=100
dodged=0
gameExit = False
while not gameExit:
for event in pygame.event.get():
if event.type == pygame.QUIT:
pygame.quit()
quit()
if event.type == pygame.KEYDOWN:
if event.key == pygame.K_LEFT:
x_change = -5
if event.key == pygame.K_RIGHT:
x_change = 5
if event.key == pygame.K_p:
pause = True
paused()
if event.type == pygame.KEYUP:
if event.key == pygame.K_LEFT or event.key == pygame.K_RIGHT:
x_change = 0
x += x_change
gameDisplay.fill(white)
things(thing_startx, thing_starty, thing_width, thing_height, black)
thing_starty+=thing_speed
car(x,y)
things_dodged(dodged)
if x > display_width - car_width or x < 0:
crash()
if thing_starty>display_height:
thing_starty = 0-thing_height
thing_startx = random.randrange(0, display_width)
dodged+=1
thing_speed+=1
#thing_width+=(dodged*1.2)
if y < thing_starty+thing_height:
print('y_crossover')
if x>thing_startx and x<thing_startx+thing_width or x+car_width>thing_startx and x+car_width<thing_startx+thing_width:
print('x crossover')
crash()
pygame.display.update()
clock.tick(60)
game_intro()
game_loop()
pygame.quit()
quit()
| 6,174 |
/machine learning_deep learning/HW copy/HW4/lec9_demo_release/.ipynb_checkpoints/train-checkpoint.ipynb | b019329a6624f0ee4c8f118fd6c418d0530c5ffb | [] | no_license | LuchaoQi/HW_JHU | https://github.com/LuchaoQi/HW_JHU | 1 | 0 | null | null | null | null | Jupyter Notebook | false | false | .py | 9,034,472 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
import re
#read file
names_file = open('names.txt', encoding = 'utf-8')
data = names_file.read()
names_file.close()
#another way to read files (wtf does 'with' mean tho)
#with open("some_file.txt") as open_file:
#data = open_file.read()
print(data)
# r means raw string saves the use of escape characters
# match only matches from the beginning of the string
last_name = r'Love'
first_name = r'Kenneth'
print(re.match(last_name,data))
print(re.match(first_name,data))
#search looks in the whole string
print(re.search(first_name,data))
# +
#challenge code
import re
file_object = open("basics.txt")
data = file_object.read()
file_object.close()
first = re.match(r'Four', data)
liberty = re.search(r'Liberty', data)
# -
# ## Escape Hatches
# \w - matches an Unicode word character. That's any letter, uppercase or lowercase, numbers, and the underscore character. In "new-releases-204",
#
# \w would match each of the letters in "new" and "releases" and the numbers 2, 0, and 4. It wouldn't match the hyphens.
#
# \W - is the opposite to \w and matches anything that isn't an Unicode word character. In "new-releases-204", \W would only match the hyphens.
#
# \s - matches whitespace, so spaces, tabs, newlines, etc.
#
# \S - matches everything that isn't whitespace.
#
# \d - is how we match any number from 0 to 9
#
# \D - matches anything that isn't a number.
#
# \b - matches word boundaries. What's a word boundary? It's the edges of word, defined by white space or the edges of the string.
#
# \B - matches anything that isn't the edges of a word.
# looking for names doesn't work bc \w matches to letters (word characters)
print(re.search(r'\w, \w', data))
#looking for phone numbers
print(re.search(r'\d\d\d-\d\d\d\d', data))
#looking for area code too, need to escape the bracket this is why raw string is important
print(re.search(r'\(\d\d\d\) \d\d\d-\d\d\d\d', data))
# ## Counts
# how to avoid writing backslash characters so many times and can use this to specify how many times a character occurs
#
# \w{3} - matches any three word characters in a row.
#
# \w{,3} - matches 0, 1, 2, or 3 word characters in a row.
#
# \w{3,} - matches 3 or more word characters in a row. There's no upper limit.
#
# \w{3, 5} - matches 3, 4, or 5 word characters in a row.
#
# \w? - matches 0 or 1 word characters.
#
# \w* - matches 0 or more word characters. Since there is no upper limit, this is, effectively, infinite word
# characters.
#
# \w+ - matches 1 or more word characters. Like *, it has no upper limit, but it has to occur at least once.
# .findall(pattern, text, flags) - Finds all non-overlapping occurrences of the pattern in the text.
# looking for names without typing a character for each letter in the name
print (re.search(r'\w+, \w+', data))
#phone numbers by using counts
print(re.search(r'\(\d{3}\) \d{3}-\d{4}', data))
# make parentesis optional by using question mark
# findall to findall phone numbers
# there is also an optional dash and space between the area code and the pnone number
print(re.findall(r'\(?\d{3}\)?-?\s?\d{3}-\d{4}', data))
#find all names
# * says either 0 up to infinite number of times
print (re.findall(r'\w*, \w+', data))
# challenge code, build function that finds all word at least inputed length long
def find_words(number,string):
return re.findall(r"\w"*number + r"\w*", string)
# ## Sets
#
# [abc] - this is a set of the characters 'a', 'b', and 'c'. It'll match any of those characters, in any order, but only once each.
#
# [a-z], [A-Z], or [a-zA-Z] - ranges that'll match any/all letters in the English alphabet in lowercase, uppercase, or both upper and lowercases.
#
# [0-9] - range that'll match any number from 0 to 9. You can change the ends to restrict the set.
#
#finding all email addresses
#set of characters can have the - , \w, \d, +, ., characters
print(re.findall(r"[-\w\d+.]+@[-\w\d.]+", data))
# all instances of "treehouse"
# ignore case flag
# re.I = re.IGNORECASE
# for a set order does not matter
print(re.findall(r"\b[trehous]{9}\b", data, re.I))
# ## Negation
# [^abc] - a set that will not match, and, in fact, exclude, the letters 'a', 'b', and 'c'.
#
# re.IGNORECASE or re.I - flag to make a search case-insensitive. re.match('A', 'apple', re.I) would find the 'a' in 'apple'.
#
# re.VERBOSE or re.X - flag that allows regular expressions to span multiple lines and contain (ignored) whitespace and comments.
# find all emails but not the ones ending in '.gov'
# re.VERBOSE for multiline strings
# use pipe | between multiple flags
# tabs are after email addresses
print(re.findall(r"""
\b@[-\w\d.]* # first a word boundary, a @ then any number of characters
[^gov\t]+ # ignore 1+ instances of the letters g,o,v and a tab
\b
""", data, re.VERBOSE | re.I))
print(re.findall(r"""
\b[-\w]+, #find a word boundary, 1+ hyphens or charascers and a coma
\s # find 1 whitespace
[-\w ]+ # 1+ hyphend and characters and excplit spaces, space in the set works
[^\t\n] # ignore tabs and newlines
""", data, re.X
))
# ## Groups
#
# ([abc]) - creates a group that contains a set for the letters 'a', 'b', and 'c'. This could be later accessed from the Match object as .group(1)
#
# (?P<name>[abc]) - creates a named group that contains a set for the letters 'a', 'b', and 'c'. This could later be accessed from the Match object as .group('name').
#
# .groups() - method to show all of the groups on a Match object.
#
# re.MULTILINE or re.M - flag to make a pattern regard lines in your text as the beginning or end of a string.
#
# ^ - specifies, in a pattern, the beginning of the string.
#
# $ - specifies, in a pattern, the end of the string.
# ^ symbol is beginnign of string $ is end of string
# mulitiline reads each line as a new string
print(re.findall(r'''
^([-\w]*,\s[-\w ]+)\t # last(optional) and first names
([-\w\d+.]+@[-\w\d.]+)\t # email
(\(?\d{3}\)?-?\s?\d{3}-\d{4})?\t # phone number(optional)
([\w\s]+,\s[\w\s.]+)\t? # Job and company
(@[\w\d]+)?$ #twitter
''', data, re.X|re.MULTILINE))
#naming groups
line = re.search(r'''
^(?P<name>[-\w]*,\s[-\w ]+)\t # last(optional) and first names
(?P<email>[-\w\d+.]+@[-\w\d.]+)\t # email
(?P<phone>\(?\d{3}\)?-?\s?\d{3}-\d{4})?\t # phone number(optional)
(?P<job>[\w\s]+,\s[\w\s.]+)\t? # Job and company
(?P<twitter>@[\w\d]+)?$ #twitter
''', data, re.X|re.MULTILINE)
print(line)
print(line.groupdict())
# +
import re
string = 'Perotto, Pier Giorgio'
names = re.match(r'(?P<last_name>\w*), (?P<first_name>\w*)', string)
print(names)
print(names.groupdict())
# +
import re
string = '''Love, Kenneth, [email protected], 555-555-5555, @kennethlove
Chalkley, Andrew, [email protected], 555-555-5556, @chalkers
McFarland, Dave, [email protected], 555-555-5557, @davemcfarland
Kesten, Joy, [email protected], 555-555-5558, @joykesten'''
contacts = re.search('(?P<email>[-\w\d+.]+@[-\w\d.]+),\s(?P<phone>\(?\d{3}\)?-?\s?\d{3}-\d{4})', string)
#use re.multiline and $ at end of regex to search end of each line
twitters = re.search(r'(?P<twitter>@[\w\d]+)$', string, re.M)
# -
# ## Complie
#
# re.compile(pattern, flags) - method to pre-compile and save a regular expression pattern, and any associated flags, for later use.
#
# .groupdict() - method to generate a dictionary from a Match object's groups. The keys will be the group names. The values will be the results of the patterns in the group.
#
# re.finditer() - method to generate an iterable from the non-overlapping matches of a regular expression. Very handy for for loops.
#
# .group() - method to access the content of a group. 0 or none is the entire match. 1 through how ever many groups you have will get that group. Or use a group's name to get it if you're using named groups.
# +
# use compile to save patterns
# add subgroups
line = re.compile(r'''
^(?P<name>[-\w]*,\s[-\w ]+)\t # last(optional) and first names
(?P<email>[-\w\d+.]+@[-\w\d.]+)\t # email
(?P<phone>\(?\d{3}\)?-?\s?\d{3}-\d{4})?\t # phone number(optional)
(?P<job>[\w\s]+,\s[\w\s.]+)\t? # Job and company
(?P<twitter>@[\w\d]+)?$ #twitter
''', re.X|re.MULTILINE)
print(re.search(line,data).groupdict())
# -
# also works same thing
print(line.search(data).groupdict())
# finditer generates iterable
for match in line.finditer(data):
print(match.group('name'))
# +
# add subgroups
line = re.compile(r'''
^(?P<name>(?P<last>[-\w]*),\s(?P<first>[-\w ]+))\t # last(optional) and first names
(?P<email>[-\w\d+.]+@[-\w\d.]+)\t # email
(?P<phone>\(?\d{3}\)?-?\s?\d{3}-\d{4})?\t # phone number(optional)
(?P<job>[\w\s]+,\s[\w\s.]+)\t? # Job and company
(?P<twitter>@[\w\d]+)?$ #twitter
''', re.X|re.MULTILINE)
# -
# NOTE ** unpacks dictionary contents to become parameters otherwise the entire dictionary will be input as one parameter
for match in line.finditer(data):
print("{first} {last} <{email}>".format(**match.groupdict()))
# +
import re
string = '''Love, Kenneth: 20
Chalkley, Andrew: 25
McFarland, Dave: 10
Kesten, Joy: 22
Stewart Pinchback, Pinckney Benton: 18'''
players = re.search(r"^(?P<last_name>[\w\s]*),\s(?P<first_name>[\w\s]*):\s(?P<score>[\d]*)$", string, re.M)
print(players)
# -
class Player():
def __init__(self, last_name, first_name, score):
self.last_name = last_name
self.first_name = first_name
self.score = score
def __str__(self):
return str(self.last_name + self.first_name + self.score)
p = Player(**players.groupdict())
print(p)
| 10,122 |
/notebooks/basic/Tutorial/2.Building_Your_Predictor.ipynb | 11be68c04e6780ba27a6d343c3de494f4e9d6cfe | [
"MIT-0"
] | permissive | transmutate/amazon-forecast-samples | https://github.com/transmutate/amazon-forecast-samples | 0 | 0 | null | 2020-04-03T07:32:35 | 2020-04-01T17:43:21 | null | Jupyter Notebook | false | false | .py | 9,870 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # SMS Spam Classifier
# [SMS Spam Collection Data Set](https://archive.ics.uci.edu/ml/datasets/SMS+Spam+Collection)
import numpy as np
import pandas as pd
from sklearn.feature_extraction.text import CountVectorizer
from sklearn.cross_validation import train_test_split
from sklearn.naive_bayes import MultinomialNB
from sklearn.feature_extraction.text import TfidfVectorizer
df = pd.read_csv('SMSSpamCollection',
sep='\t',
names=['Status', 'Message'])
df.head()
# Total number of examples
len(df)
# Number of spam examples
len(df[df.Status=='spam'])
# Replace ham with 0 and spam with 1
df.loc[df["Status"] == 'ham', "Status"] = 0
df.loc[df["Status"] == 'spam', "Status"] = 1
df.head()
# Split the data
df_x = df["Message"]
df_y = df["Status"]
vectorizer = CountVectorizer(stop_words = 'english')
#vectorizer = TfidfVectorizer(stop_words = 'english')
x_train, x_test, y_train, y_test = train_test_split(df_x, df_y, test_size=0.2, random_state=4)
features = vectorizer.fit_transform(x_train)
words_array = features.toarray()
words_array
words_array[0]
len(words_array[0])
vectorizer.inverse_transform(words_array[0])
x_train.iloc[0]
mnb = MultinomialNB()
y_train = y_train.astype('int') # Cast y_train to integer values
mnb.fit(features, y_train)
test_features = vectorizer.transform(x_test)
predictions = mnb.predict(test_features)
predictions
actual = np.array(y_test)
actual
num_correct = (predictions == actual).sum()
num_correct
total_examples = len(predictions)
total_examples
# Get the accuracy
accuracy = num_correct / total_examples * 100
round(accuracy, 2)
mnb.predict(features[0])
x_test.iloc[0]
| 1,940 |
/Sismologia/Exercícios - Pontenciais Sísmicos- Sismologia.ipynb | 6e7a5e6586ae90a91746d8cfe933bd95182b16fa | [] | no_license | dIOGOLOC/Projeto-Doutorado | https://github.com/dIOGOLOC/Projeto-Doutorado | 1 | 0 | null | null | null | null | Jupyter Notebook | false | false | .py | 11,364 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# 1. [신경망] 다음 코드를 무엇을 의미하는지 이해하고 실행하여 결과를 확인하세요.(14점)
# (코드의 해석과 결과의 의미를 작성하세요.)
# +
# torch.nn 패키지를 사용하여 신경망을 생성함.
# nn 패키지는 모델을 정의할 때, autograd를 통해 자동미분 기능을 제공함
# nn.Module은 층과 전방전파(forward propagation) (입력 -> 출력)을 쉽게 구현함
# 참고로 nn 패키지는 mini-batch만 지원함,
# 예로 nn.Conv2d는 4차 Tensor를 받음(nSamples*nChannels*height*width)
# 아래 AlexNet(이미 수업에서 학습함.) 예시는 숫자를 분류하는 간단한 컨볼루션 신경망의 예임
# -
# 
# +
# 예시는 사진 입력을 받고, 몇 개의 층에 전방 전파하면서 분류를 출력함
# 출력을 위해서 모델은 다음과 같은 학습을 수행함
# - 신경망은 학습가능한 매개변수들(가중치들)을 가짐
# - 사진 데이터를 반복적으로 입력함
# - 신경망을 통해 입력을 처리함(전방 전파)
# - 손실(오차)를 계산함(실제 출력과 예측 출력을 비교하여 학습의 올바름을 판단함)
# - 오차로부터 그레이디언트(경사, 방향)을 신경망의 각 매개변수에게 역전파함(오류 역전파)
# - 신경망의 매개변수들을 갱신함((미래)가중치 = (현재)가중치 - 학습률 * 그레이디어트)
# 위의 컨볼루션 신경망의 부분들을 torch를 통해서 손쉽게 구현할 수 있음.
# 단지 forward 함수만 정의하면, autograd를 이용해서
# 해당 연산 그래프의 그레이디언트를 구하는 backward 자동적으로 정의됨.
# forward 함수는 Tensor를 이용할 수 있는 다양한 연산들(합, 곱 등등) 사용하여 정의 가능함.
# torch.Tensor: 자동 미분 기능을 지원하는 다차원 배열, 각 Tensor에 해당하는 그레이디언트를 가짐.
# nn.Module: 신경망 모듈이며 매개변수의 캡슐화, GPU 연산 등 작업을 쉽게 가능하게 함.
# nn.Parmeter: 모듈이 지정되면 매개변수들을 자동으로 관리하는 Tensor의 하나임.
import torch
import torch.nn as nn
import torch.nn.functional as F
class Net(nn.Module):
def __init__(self):
# for parent class
super(Net, self).__init__()
# 1 input image channel, 6 output channels, 5x5 square convolution
# kernel(filter)
self.conv1 = nn.Conv2d(1, 6, 5)
# 6 input image channer, 16 output channels, 5x5 square convolution
self.conv2 = nn.Conv2d(6, 16, 5)
# an affine operation: y = Wx + b
self.fc1 = nn.Linear(16 * 5 * 5, 120)
self.fc2 = nn.Linear(120, 84)
self.fc3 = nn.Linear(84, 10)
def forward(self, x):
# Max pooling over a (2, 2) window
x = F.max_pool2d(F.relu(self.conv1(x)), (2, 2))
# If the size is a square you can only specify a single number
x = F.max_pool2d(F.relu(self.conv2(x)), 2)
x = x.view(-1, self.num_flat_features(x)) # reshape in tensorflow
x = F.relu(self.fc1(x))
x = F.relu(self.fc2(x))
x = self.fc3(x) # output layer
return x
def num_flat_features(self, x):
size = x.size()[1:] # all dimensions except the batch dimension
num_features = 1
for s in size:
num_features *= s
return num_features
net = Net()
print(net)
# (1) 화면 출력 확인 및 의미를 서술
# -
# 위의 출력은 Network 구성을 보여준다.
#
# - convolution1(1 = input depth, output depth = 6, kernel = 5 x 5, stride=1)
# - convolution2(6 = input depth, output depth = 16, kernel = 5 x 5, stride=1)
# - fully-connected layer(input_node = 400, output_node=120, including=bias term)
# - fully-connected layer(input_node = 120, output_node=84, including=bias term)
# - fully-connected layer(input_node = 84, output_node=10, including=bias term)
#
# 물론 각 layer 마다 activation function으로 ReLU(rectified Linear unit)을 사용하는
# CNN의 구조를 보여준다.(마지막 output layer에서는 activation function을 사용하지 않는다.)
# +
# (2) 정의된 컨볼루션 신경망의 구조 설명 (위의 AlexNet 그림 참고)
# -
# 전체적인 구조는
#
# - convolution1(1 = input depth, output depth = 6, kernel = 5 x 5, stride=1)
# - max_pooling(convolution1) : subsampling
# - convolution2(6 = input depth, output depth = 16, kernel = 5 x 5, stride=1)
# - max_pooling(convolution2) : subsampling
# - fully-connected layer(input_node = 400, output_node=120, including=bias term)
# - fully-connected layer(input_node = 120, output_node=84, including=bias term)
# - fully-connected layer(input_node = 84, output_node=10, including=bias term)
#
# 물론 각 layer 마다 activation function으로 ReLU(rectified Linear unit)을 사용하는
# CNN의 구조를 보여준다.(마지막 output layer와 max pooling에서는 activation function을 사용하지 않는다.)
# 위와 코드는 위와 같은 구조로 구성되어진다.
# net.parameters()를 사용하여 정의된 신경망의 학습가능한 매개변수들을 확인할 수 있음
params = list(net.parameters())
print(len(params))
print(params[0].size()) # conv1's .weight
# (3) 화면 출력 확인
# 위에서 설계한 convolution network의 매개변수 수와
# first conv layer의 weight size를 출력을 하고 있다.
# 다음의 임의의 32*32 입력을 가정함
# 참고로 크기가 다른 입력을 받을 때는 입력의 크기를 재조정하거나 신경망 수정함
# 입력 이미지 크기 batch, channel, height, width
input = torch.randn(1, 1, 32, 32)
out = net(input)
print(out)
# (4)화면 출력 확인
# 최종 10개의 class의 분류 score를 보여주고 있다.
# +
# 오류역전파를 통해 그레이디언트를 구하기 전에 모든 가중치의 그레이디언트 버퍼들을 초기화
net.zero_grad()
# in backward, compute gradient w.r.t torch.randn(1,10) tensor
out.backward(torch.randn(1,10))
# 손실 함수 정의 및 임의의 값들에 대해서 오차 결과 확인
# nn 패키지는 많이 사용되는 손실함수들을 제공하며, 해당 예제는 단순한 MSE를 사용
output = net(input)
target = torch.randn(10) # a dummy target, for example
print(target)
target = target.view(1, -1) # make it the same shape as output
print(target)
criterion = nn.MSELoss()
loss = criterion(output, target)
print("Checking loss value")
print(loss)
# (5) 화면 출력 확인
# +
# 앞에 코드에서 언급한 것과 같이 오류 역전파하기 전, 그레이디언트를 초기화해야 함
# backward() 수행 후 어떤 변화가 있는지 확인하고, 초기화의 필요성을 확인함
net.zero_grad() # zeroes the gradient buffers of all parameters
print("conv1.bias.grad before backward")
print(net.conv1.bias.grad)
# (6) 화면 출력 확인
# 오류 역전파하기전의 conv1.bias의 gradient를 값을 보여준다.
# 오류 역전파하기전인 만큼 0의 값으로 초기화 되어진다.
# +
# loss에 기반하여 backward를 실행한다.
loss.backward()
print("conv1.bias.grad after backward")
print(net.conv1.bias.grad)
# (7) 화면 출력 확인
# 오류 역전파가 수행이 되면서
# conv1.bias의 gradient 값을 보여준다.
# +
# 스토캐스틱경사하강법((미래)가중치 = (현재)가중치 - 학습률* 그레이디언트)을
# 이용하여 가중치 갱신하는 코드는 다음과 같음
learning_rate = 0.01
for f in net.parameters():
f.data.sub_(f.grad.data * learning_rate)
# 하지만 위 구현 코드보다 실제, torch.optim에서 구현되는 SGD, Adam, RMSProp 등을 사용함
# 오류 역전파에서 최적화하는 방법을 보인 예제 코드
import torch.optim as optim
# create your optimizer
optimizer = optim.SGD(net.parameters(), lr=0.01)
# in your training loop:
optimizer.zero_grad() # zero the gradient buffers
output = net(input)
loss = criterion(output, target)
loss.backward()
optimizer.step() # Does the update
# -
# 2. [분류기 학습] 다음 코드를 무엇을 의미하는지 이해하고 실행하여 결과를 확인하세요.(14점)
# (코드의 해석과 결과의 의미를 작성하세요.)
# +
# 1번의 기초적인 신경망을 통해서 사진 분류기를 학습함
# 데이터집합은 CIFAF-10이며,
# 아래의 예와 같이 10가지의 3(R, G, B) 채넝릐 32*32 크기의 사진으로 구성됨
# -
# 
# +
# CIFAR-10과 같이 많이 사용되는 데이터집합은 torchvision 패키지에서 제공함
# 분류기 학습은 다음과 같은 과정을 가짐
# 1. 정규화된 CIFAR-10 훈련집합과 테스트집합을 torchvision을 이용하여 적재함
# 2. 컨볼루션 신경망을 정의함
# 3. 손실함수 정의
# 4. 훈련집합을 이용하여 신경망을 학습시킴
# 5. 테스트집합을 이용하여 신경망 성능 확인
### 1. 정규화된 CIFAR-10 훈련집합과 데스트 집합을 torchvision을 이용하여 적재함
import torch
import torchvision
import torchvision.transforms as transforms
# transforms.Normalize function normalize a tensor image with -
# mean and standard deviation channel by channel
transform = transforms.Compose(
[transforms.ToTensor(),
transforms.Normalize((0.5, 0.5, 0.5), (0.5, 0.5, 0.5))])
trainset = torchvision.datasets.CIFAR10(root='./data', train=True,
download=True, transform=transform)
trainloader = torch.utils.data.DataLoader(trainset, batch_size=4,
shuffle=True, num_workers=2)
testset = torchvision.datasets.CIFAR10(root='./data', train=False,
download=True, transform=transform)
testloader = torch.utils.data.DataLoader(testset, batch_size=4,
shuffle=False, num_workers=2)
classes = ('plane', 'car', 'bird', 'cat',
'deer', 'dog', 'frog', 'horse', 'ship', 'truck')
# (1) 화면 출력 확인
print("trian set: {}".format(len(trainset)))
print("test set: {}".format(len(testset)))
# +
# 훈련집합의 일부 사진들 확인
import matplotlib.pyplot as plt
import numpy as np
# functions to show an image
def imshow(img):
img = img / 2 + 0.5 # unnormalize
npimg = img.numpy()
plt.imshow(np.transpose(npimg, (1, 2, 0)))
# get some random training images
dataiter = iter(trainloader)
images, labels = dataiter.next()
print("The size of image tensor:", images.size())
print("The size of label tensor:", labels.size(), labels[0])
# show images
imshow(torchvision.utils.make_grid(images))
# print labels
print(' '.join('%5s' % classes[labels[j]] for j in range(4)))
# (2) 화면 출력 확인
# +
### 2. 컨볼루션 신경망을 정의함
# 3 채널 32*32 크기의 사진을 입력받고, 신경망을 통과해 10분류를 수행
import torch.nn as nn
import torch.nn.functional as F
class Net(nn.Module):
def __init__(self):
super(Net, self).__init__()
self.conv1 = nn.Conv2d(3,6,5)
self.pool = nn.MaxPool2d(2,2)
self.conv2 = nn.Conv2d(6, 16, 5)
self.fc1 = nn.Linear(16*5*5, 120)
self.fc2 = nn.Linear(120, 84)
self.fc3 = nn.Linear(84, 10)
def forward(self, x):
x = self.pool(F.relu(self.conv1(x)))
x = self.pool(F.relu(self.conv2(x)))
x = x.view(-1, 16*5*5)
x = F.relu(self.fc1(x))
x = F.relu(self.fc2(x))
x = self.fc3(x)
return x
net = Net()
### 3. 손실함수 정의, 교차 엔트로피와 SGD + Momentum
import torch.optim as optim
criterion = nn.CrossEntropyLoss()
optimizer = optim.SGD(net.parameters(), lr=0.001, momentum=0.9)
### 4. 훈련집합을 이용하여 신경망을 학습시킴
for epoch in range(2): # loop over the dataset multiple times
running_loss = 0.0
for i, data in enumerate(trainloader, 0):
# Get the inputs
inputs, labels = data
# zero the parameter gradients
optimizer.zero_grad()
# Forward + Backward + Optimize
outputs = net(inputs)
loss = criterion(outputs, labels)
loss.backward()
optimizer.step()
# print statistics
running_loss += loss.item()
if i % 1000 == 999: # print every 1000 mini-batches
print("[%d, %5d]loss: %.3f" % (epoch + 1, i +1, running_loss / 1000))
running_loss = 0.0
print("Finished training")
# (3) 화면 출력 확인 및 학습이 되고 있는지 서술
# -
# 위의 loss 값을 확인해보면 점차 낮아지는 방향으로 학습이 진행이 되고 있다.
# +
### 5. 테스트 집합을 이용하여 신경망 성능 확인
dataiter = iter(testloader)
images, labels = dataiter.next()
# show images
imshow(torchvision.utils.make_grid(images))
# print labels
print("GroundTruth:", ' '.join('%5s' % classes[labels[j]] for j in range(4)))
# (4) 화면 출력 확인
# -
outputs = net(images)
_, predicted = torch.max(outputs, 1)
print("Predicted:", ' '.join('%5s' % classes[predicted[j]] for j in range(4)))
# (5) 화면 출력 확인
# 예측된 label의 type을 출력하고 있다.
print("예측된 label의 type을 출력하고 있다. ")
# +
# Perfomance on the whole test dataset
correct = 0
total = 0
with torch.no_grad():
for data in testloader:
images, labels = data
output = net(images)
_, predicted = torch.max(output.data, 1)
total += labels.size(0)
correct += (predicted == labels).sum().item()
print('Accuracy of the network on the 10000 test images: %d %%' %
(100 * correct/total))
# (6) 화면 출력 확인 및 일반화 성능 서술
# -
# 위의 결과를 보면 알 수 있듯 일반화 성능은 testset을 가지고 나타낼 수 있는데
# 이것은 training을 할 때 사용하지 않은 데이터를 가지고 나의 모델이 training되지 않은
# 데이터를 얼마나 잘 예측을 할 수 있는지 수치를 정확도로 판단하는데 현재 모델은
# 정확도(accuracy) 54%를 보여준다.
# +
# Performance on each class
class_correct = list(0. for i in range(10))
class_total = list(0. for i in range(10))
print("The initial class_correct:", class_correct)
print("The initial class_total:", class_total)
with torch.no_grad():
for data in testloader:
images, labels = data
outputs = net(images)
_, predicted = torch.max(output, 1)
c = (predicted == labels).squeeze()
for i in range(4):
label = labels[i]
class_correct[label] += c[i].item()
class_total[label] += 1
for i in range(10):
print('Accuracty of %5s: %2d %%' % (
classes[i], 100* class_correct[i]/class_total[i]))
# (7) 화면 출력 확인 및 부류별 분류기의 성능 서술
# -
# 위의 결과는 10개의 class 마다의 recall이 얼마나 잘된 결과인지를 보여준다.
# 우선 출력 결과를 보면, cat(24%), dog(25%), horse(25%), truck(25%)의 recall을
# 보여주고 나머지의 경우는 0%의 recall을 보여주고 있습니다.
# 3. 다음 조건을 만족하는 컨볼루션 신경망을 구현하고, 2번의 (3), (6), (7)의 결과를 확인하고 비교하세요.
#
# (1) INPUT-CONV(32 3\*3)-CONV(32 3\*3)-RELU-POOL-CONV(32 3\*3)-CONV(32 3\*3)-RELU-POOL-FC-OUTPUT(15점)
#
# (2) 2번 문제의 신경망에 Adam 최적화(강의자료의 기본 하이퍼 매개변수 사용) 적용(3점)
#
# (3) 데이터 확대 방법들 중 하나늘 적용한 후, 2 번 문제의 신경망 학습(Hint: transforms) (3점)
#
# (4) 2번 문제의 신경망에 CONV 층마다 배치 정규화를 적교(Hint: nn.BatchNorm) (3점)
#
# (5) 2번 문제의 신경망에 로그우드 손실함수를 적 (3점)
#
# (6) 2번 문제의 신경망에 L2 Norm 적용(3점)
# +
### 1. 정규화된 CIFAR-10 훈련집합과 데스트 집합을 torchvision을 이용하여 적재함
import torch
import torchvision
import torchvision.transforms as transforms
# transforms.Normalize function normalize a tensor image with -
# mean and standard deviation channel by channel
# (3) 데이터 확대 방법들 중 하나늘 적용한 후, 2 번 문제의 신경망 학습(Hint: transforms) (3점)
transform = transforms.Compose(
[transforms.Resize(128),
transforms.CenterCrop(32),
transforms.ToTensor(),
transforms.Normalize((0.5, 0.5, 0.5), (0.5, 0.5, 0.5))])
trainset = torchvision.datasets.CIFAR10(root='./data', train=True,
download=True, transform=transform)
trainloader = torch.utils.data.DataLoader(trainset, batch_size=4,
shuffle=True, num_workers=2)
testset = torchvision.datasets.CIFAR10(root='./data', train=False,
download=True, transform=transform)
testloader = torch.utils.data.DataLoader(testset, batch_size=4,
shuffle=False, num_workers=2)
classes = ('plane', 'car', 'bird', 'cat',
'deer', 'dog', 'frog', 'horse', 'ship', 'truck')
# (1) 화면 출력 확인
print("trian set: {}".format(len(trainset)))
print("test set: {}".format(len(testset)))
### 2. 컨볼루션 신경망을 정의함
# 3 채널 32*32 크기의 사진을 입력받고, 신경망을 통과해 10분류를 수행
import torch.nn as nn
import torch.nn.functional as F
# (1) INPUT-CONV(32 3*3)-CONV(32 3*3)-RELU-POOL-
# CONV(32 3*3)-CONV(32 3*3)-RELU-POOL-FC-OUTPUT(15점)
class Net2(nn.Module):
def __init__(self):
super(Net2, self).__init__()
self.conv1 = nn.Conv2d(3,32,3)
# (4) 2번 문제의 신경망에 CONV 층마다 배치 정규화를 적교(Hint: nn.BatchNorm) (3점)
self.batch_norm1 = nn.BatchNorm2d(32)
self.conv2 = nn.Conv2d(32, 32, 3)
self.pool1 = nn.MaxPool2d(2,2)
self.batch_norm2 = nn.BatchNorm2d(32)
self.conv3 = nn.Conv2d(32, 32, 3)
self.batch_norm3 = nn.BatchNorm2d(32)
self.conv4 = nn.Conv2d(32, 32, 3)
self.pool2 = nn.MaxPool2d(2,2)
self.fc1 = nn.Linear(32*5*5, 10)
def forward(self, x):
x = self.conv1(x)
#print(x.size())
x = self.batch_norm1(x)
#print(x.size())
x = self.pool1(F.relu(self.conv2(x)))
#print(x.size())
x = self.batch_norm2(x)
x = self.conv3(x)
x = self.batch_norm3(x)
#print(x.size())
x = self.pool2(F.relu(self.conv4(x)))
#print(x.size())
x = x.view(-1, 32*5*5)
x = self.fc1(x)
return x
net = Net2()
### 3. 손실함수 정의, Loglikelihood + Adam
import torch.optim as optim
# (2) 2번 문제의 신경망에 Adam 최적화(강의자료의 기본 하이퍼 매개변수 사용) 적용(3점)
# (6) 2번 문제의 신경망에 L2 Norm 적용(3점) - weight_decay=1e-05
optimizer = optim.Adam(net.parameters(), lr = 0.001,
betas=(0.9, 0.999), weight_decay=1e-05)
### 4. 훈련집합을 이용하여 신경망을 학습시킴
for epoch in range(2): # loop over the dataset multiple times
running_loss = 0.0
for i, data in enumerate(trainloader, 0):
# Get the inputs
inputs, labels = data
if i == 0 and epoch == 0:
print("The size of inputs tensor:", inputs.size())
print("The size of labels tensor:", labels.size())
# zero the parameter gradients
optimizer.zero_grad()
# Forward + Backward + Optimize
outputs = net(inputs)
# (5) 2번 문제의 신경망에 로그우드 손실함수를 적 (3점)
loss = F.nll_loss(F.log_softmax(outputs), labels)
loss.backward()
optimizer.step()
# print statistics
running_loss += loss.item()
if i % 1000 == 999: # print every 1000 mini-batches
print("[%d, %5d]loss: %.3f" % (epoch + 1, i +1, running_loss / 1000))
running_loss = 0.0
print("Finished training")
# -
# 4. 신경망의 출력이 (0.4, 2.0, 0.001, 0.32).T 일 때, 소프트 맥스 함수를 적용한 결과를 쓰시오. (6점)
# +
import numpy as np
output = np.array([0.4, 2.0, 0.001, 0.32])
#print(output)
def print_softmax(e):
print("The softmax of each rows")
for i in range(len(e)):
print(i, e[i])
print("The total of softmax: {}".format(np.sum(e)))
def softmax_funtion(x):
exp = np.exp(output)
#print(exp)
softmax_val = exp / np.sum(exp)
print_softmax(softmax_val)
softmax_funtion(output)
# -
# 5. 소프트맥스 함수를 적용한 후 출력이 (0.001, 0.9, 0.001, 0.098).T 이고 레이블 정보가 (0, 0, 0, 1).T 일때, 세 가지 목적함수, 평균제곱 오차, 교차 엔트로피, 로그우드를 계산하시오.
# +
import numpy as np
output = np.array([0.001, 0.9, 0.001, 0.098])
ground_truth = np.array([0, 0, 0, 1])
def mean_squared_error(out, truth):
error = np.mean((out - truth)**2)
return error
def cross_entroy(out, truth):
error = truth*np.log(output)
return np.sum(error)
def loglikelihood(out, index, base=True):
if base == True: # base 2
error = np.log2(out[index])
else: # base e
error = np.log(out[index])
return error
print("MSE(mean squared error): {}".format(mean_squared_error(output, ground_truth)))
print()
print("cross_entroy: {}".format(mean_squared_error(output, ground_truth)))
print()
print("loglikelihood: {}".format(mean_squared_error(output, 3)))
# -
# # Reference
#
#
# - [pytorch](https://pytorch.org/docs/stable/nn.html?highlight=nll#torch.nn.functional.nll_loss)
#
# - [Getting started with PyTorch for Deep Learning](https://codetolight.wordpress.com/2017/11/29/getting-started-with-pytorch-for-deep-learning-part-3-neural-network-basics/)
#
# - [data augment of pytorch in stackoverflow](https://stackoverflow.com/questions/50002543/transforms-compose-meaning-pytorch)
#
# - [L2 regularization in stackoverflow](https://stackoverflow.com/questions/42704283/adding-l1-l2-regularization-in-pytorch)
#
| 18,192 |
/Python/Python-for-Algorithms--Data-Structures--and-Interviews/12 Array Sequences/Array Sequence Interview Questions/.ipynb_checkpoints/Sentence Reversal-checkpoint.ipynb | 227cd56d57809ef4cdd15cfa450c39423e99bc06 | [] | no_license | cowboyuniverse/LearningJournal | https://github.com/cowboyuniverse/LearningJournal | 0 | 1 | null | null | null | null | Jupyter Notebook | false | false | .py | 9,149 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Python 2
# language: python
# name: python2
# ---
# # Sentence Reversal
#
# ## Problem
#
# Given a string of words, reverse all the words. For example:
#
# Given:
#
# 'This is the best'
#
# Return:
#
# 'best the is This'
#
# As part of this exercise you should remove all leading and trailing whitespace. So that inputs such as:
#
# ' space here' and 'space here '
#
# both become:
#
# 'here space'
#
# ## Solution
#
# Fill out your solution below:
# +
def rev_word(s):
words = []
chars = []
for char in s:
if char is " " and chars:
words.append("".join(chars))
chars =[]
elif char is not " ":
chars.append(char)
if chars:
words.append("".join(chars))
words = revserse(words)
return words
def revserse(s):
sentence = ""
length = len(s) - 1
while length >= 0:
sentence += "".join(s[length]) + " "
length-=1
return sentence
# -
rev_word('Hi John, are you ready to go?')
rev_word(' space before')
# _____
# # Test Your Solution
# +
"""
RUN THIS CELL TO TEST YOUR SOLUTION
"""
from nose.tools import assert_equal
class ReversalTest(object):
def test(self,sol):
assert_equal(sol(' space before'),'before space')
assert_equal(sol('space after '),'after space')
assert_equal(sol(' Hello John how are you '),'you are how John Hello')
assert_equal(sol('1'),'1')
print "ALL TEST CASES PASSED"
# Run and test
t = ReversalTest()
t.test(rev_word)
# -
# ## Good Job!
ści ustawiamy ziarno generatora pseudolosowego, aby w każdym uruchomieniu wyniki były takie same.
np.random.seed(2)
# Najpierw generuje losowy ciąg bitów o długości powiedzmy 100:
Alice_bits=np.random.randint(0,2,100)
Alice_bits
# Następnie musi wygenerować losowy ciąg baz, zatem generuje znowu losowy ciąg bitów, i przyjmuje, że $0$ odpowiada bazie $Z$ a $1$ bazie $X$. Oba losowe ciągi Alicja, póki co, zachowuje wyłacznie dla siebie.
#wylosuj bazy
Alice_bases=np.random.randint(0,2,100)
Alice_bases
# Ciag bitów wygląda tak: [0,1,1,...] a ciąg baz tak [0,1,0,..] oznacza to, ze pierwszy foton (nasz kubit) Alicja koduje w bazie $0$ czyli w bazie $Z$ i przypisuje mu bit $0$ czyli stan $|0\rangle$, drugi foton koduje w bazie $1$ czyli $X$ przypisuje mu bit $1$ czyli stan $|-\rangle$, trzeci foton w bazie $0$ czyli $Z$ przypsiuje mu bit $1$ czyli stan $|1\rangle$, itd. Musimy zatem stworzyć obwody kwantowe generujące odpowiednie stany. W rzeczywistości byłyby to fotony a ich stany byłyby ustawiane za pomocą polaryzatora, my robimy to na komputerze kwantowym (a właściwie na symulatorze), zamisat fotonów mamy kubity i zmieniamy ich stan aplikując odpowiednie bramki. Napisz funkcję, która jako argumenty przyjmuje listy $bits$ i $bases$, a zwraca listę obwodów kwantowych generujących odpowiednie stany, nie dodawaj pomiaru na końcu obwodu, dodaj na końcu barrierę dla lepszej wizualizacji poszczególnych etapów, oraz dodaj jeden bit klasyczny, który bedzie potrzebny do późniejszych pomiarów.
def prepare_states(bits,bases):
w = []
for i in range(bits.size):
w.append(QuantumCircuit(1,1))
if bits[i] == 0 and bases[i] == 0:
w[i].barrier()
#print(0)
if bits[i] == 1 and bases[i] == 0:
w[i].x(0)
w[i].barrier()
#print(1)
if bits[i] == 0 and bases[i] == 1:
w[i].h(0)
w[i].barrier()
#print("+")
if bits[i] == 1 and bases[i] == 1:
w[i].x(0)
w[i].h(0)
w[i].barrier()
#print("-")
return w
# Alicja generuje stany:
states=prepare_states(Alice_bits,Alice_bases)
# Tak powinno wyglądać trzy pierwsze stany z listy:
states[0].draw(output='mpl')
states[1].draw(output='mpl')
states[2].draw(output='mpl')
# Następnie Alicja powinna wysłać tak przygotowane fotony Bobowi, w naszym przypadku mamy do dyspozycji kubity na stałe osadzone w komputerze kwantowym zatem zakladamy, że teraz przejął je Bob i wykonuje na nich operacje. Bob na otrzymanych od Alicji kubitach dokonuje pomiaru, jednak nie wie w jakiej bazie dokonać pomiaru bo Alicja używała losowo wybranych baz. Zatem Bob losuje bazy w których będzie dokonywał pomiaru (oczywiście dostanie inny ciąg baz niż Alicja ale jak się przekonamy to nic nie szkodzi).
#wylosuj bazy Boba
Bob_bases=np.random.randint(0,2,100)
# Aby dokonać pomiaru w bazie $Z$ nie musimy nic robić, po prostu dokonujemy pomiaru na kubicie, wynik $0$ odpowiada stanowi $|0\rangle$ a wynik $1$ stanowi $|1\rangle$. Aby dokonać pomairu w stanie $X$ musimy najpierw obrócić odpowiednio stan za pomocą bramki Hadamarda i dopiero potem dokonać pomiaru, wtedy wynik $0$ będzie odpowiadał stanowi $|+\rangle$ a wynik $1$ stanowi $|-\rangle$. Napisz funkcję, która jako argumenty przyjmuje stany $states$ otrzymane przez Boba od Alicji i wylosowane przez Boba bazy $Bob\_bases$ a zwraca bity bądące wynikami pomiarów Boba. Użyj symulatoraz pojedyńczym wykonaniem obwodu:
#
# backend = BasicAer.get_backend('qasm_simulator')
#
# execute(qc, backend, shots=1).result().get_counts()
def measure_states(states,bases):
backend = BasicAer.get_backend('qasm_simulator')
w = []
for i in range(bases.size):
qc = states[i]
if(bases[i] == 1):
qc.h(0)
qc.measure([0],[0])
c = execute(qc, backend, shots=1).result().get_counts()
if c == {'0': 1}:
w.append(0)
if c == {'1': 1}:
w.append(1)
return(w)
Bob_bits=measure_states(states,Bob_bases)
# Tutaj wyniki bedą losowe, więc ich nie porównujemy. Zastanówmy się co się stało wyniku tych pomiarów. Zarówno Alicja jak i Bob używali losowo wybranych baz, zatem w niektórych przypadkach baza Alicji i Boba będzie taka sama a w niektórych nie. W przypadku gdy bazy są takie same Bob z pomiaru odczyta z prawdopodobieństwe równym 1 bit taki jaki zakodowała Alicja, tzn. jeżeli np. Alicja użyła bazy $X$ i zakodowała bit $0$ czyli wysłała stan $|+\rangle$ to Bob używając bazy $X$ do pomiaru z pradopodobieństwem równym jeden otrzyma wynik $0$ odpowiadający stanowi $|+\rangle$, jeżeli natomiast Bob użyje błędnej bazy to z prawdopodobieństwem $\frac{1}{2}$ odczyta poprawny bit a z prawdopobieństwem $\frac{1}{2}$ błędny. Jeżeli np. Alicja użyła bazy $X$ i zakodowała bit $0$ czyli wysłała stan $|+\rangle$ to Bob używając bazy $Z$ do pomiaru z prawdopodobieństwem równym $\frac{1}{2}$ otrzyma wynik $0$ odpowiadający stanowi $|+\rangle$ i z prawdopodobieństwem równym $\frac{1}{2}$ otrzyma wynik $1$ odpowiadający stanowi $|-\rangle$.
#
# Następnie Alicja ujawnia bazy w których kodowały stany, czyli ujawnia ciąg $Alice\_bases$:
Alice_bases
# Alicja i Bob mogą teraz porównać swój wybór baz $Bob\_bases$ i $Alice\_bases$ i odrzucić ze swoich wyników te bity na których ich bazy nie są ze sobą zgodne. Napsiz funkcję, która jako argumenty przyjmuje bazy Boba i Alicji oraz bity kodowane przez Alicję $Alce\_bits$ oraz zmierzone przez Boba $Bob\_bits$ i zwraca dwa ciągi bitów, jeden Alicji a drugi Boba, które odpowiadają przypadkom kiedy bazy Alicji i Boba się zgadzały.
def common_bits(Alice_bases,Bob_bases,Alice_bits,Bob_bits):
ak = []
bk = []
for i in range(Alice_bases.size):
if(Alice_bases[i] == Bob_bases[i]):
ak.append(Alice_bits[i])
bk.append(Bob_bits[i])
return ak,bk
Alice_key, Bob_key=common_bits(Alice_bases,Bob_bases,Alice_bits,Bob_bits)
# W ten sposób Alicja i Bob otrzymali pewien klucz, który powinien być wspólny dla nich obu, o ile nie było podsłuchu po drodze (o tym co wtedy za chwilę). Aby Alicja i Bob sprawdzili czy nie było podsłuchu muszą ujawnić jakiś fragment swoich kluczy i go porównać, powiedzmy 15 pierwszych bitów.
# +
print(Alice_key)
print(Bob_key)
Alice_key[:15]==Bob_key[:15]
# -
# W ten sposób mają (z pewnym prawdopodobieństwem) przekonanie, że ich klucze są jednakowe i nikt inny ich nie zna. Możemy jeszcze sprawdzić, że rzeczywiście ich klucze są w całości jednakowe:
Alice_key==Bob_key
# Oczywiście w rzeczywistości Alcija i Bob tego nie robią bo musieliby ujawnić całe swoje klucze. Spójrzmy jeszcze na długość uzyskanych kluczy, powinna wynosić ok. 50. Wynika to z tego, że mieliśmy 100 fotonów a Bob miał 0.5 szansy na wylosowanie dobrej bazy.
len(Alice_key)
len(Bob_key)
# W ten sposób Alicja i Bob mają wspólny ciąg bitów. Zastanówmy się teraz co z bezpieczeństwem takiego protokołu. Załóżmy, że w naszym układzie pojawia się trzecia osoba, Ewa, która chce poznać klucz uzgadniany przez Alicję i Boba. Ewa przechwytuje fotony (kubity) przesyłane przez Alicję do Boba i dokonuje na nich pomiaru, oczywiscie Ewa nie zna ciągu baz w których Alicja zakodowała bity, bo Alicja trzyma je w sekrecia tak długo aż fotony nie dotrą do Boba. Zatem Ewa musi dokonywać pomiaru w losowych bazach. Ewa otrzyma częśc wyników poprawnych a częśc nie ale jednocześnie zniszczy stany kwantowe fotonów. Jak pamiętamy z poprzedniej listy, w wyniku pomiaru stan kwantowy kolapsuje, zatem Ewa dokonjąc losowych pomiarów sprawi, że skolapsują one w losowy sposób. Ewa po dokonaniu pomiarów odsyła fotony dalej do Boba. Ten dokonuje znowu losowych pomiarów jak przedtem, ale tym razem część stanów na których dokonuje pomiaru będzie zmieniona przez pomiar dokonany przez Ewę, zatem nawet tam gdzie Bob użyje do pomiaru tej samej bazy co Alicja, będzie mógł dostać błędny wynik, z tego powodu, że stan wcześniej zmodyfikowała Ewa. Zatem aby się przekonać czy Ewa podsłuchiwała Alicja i Bob muszą ujawnić część swoich uzgodnionych bitów (powiedzmy 15) i sprawdzić czy się one zgadzają ze sobą.
Alice_key[:15]==Bob_key[:15]
# Bity się zgadzają zatem jest wysokie prawdopodobieństwo, że Ewa nie podsłuchiwała i pozostałe bity mogą posłużyć jako wspólny tajny klucz. Zobaczmy teraz co sie dzieje gdy Ewa podsłuchuje. Zacznijmy protokół od nowa, Alicja generuje stany:
Alice_bits=np.random.randint(0,2,100)
Alice_bases=np.random.randint(0,2,100)
states=prepare_states(Alice_bits,Alice_bases)
# Następnie przechwytuje je Ewa i póbuje odczytać informacje:
Eve_bases=np.random.randint(0,2,100)
Eve_bits=measure_states(states,Eve_bases)
# Potem odsyła je do Boba, pamiętajmy jednak, że w wyniku pomiaru Ewy częśc z nich została zmieniona. Teraz Bob dokonuje pomiaru.
Bob_bases=np.random.randint(0,2,100)
Bob_bits=measure_states(states,Bob_bases)
# teraz Alicja i Bob porównują swoje bazy:
Alice_key, Bob_key=common_bits(Alice_bases,Bob_bases,Alice_bits,Bob_bits)
# następnie ujawniają część ze swoich bitów, jeżeli któreś się nie zgadzają to znaczy, ze Ewa podsłuchiwała.
Alice_key[:15]==Bob_key[:15]
# w takiej sytuacji Alicja i Bob odrzucają wszystkie bity jako, że były podsłuchiwane.
# ## Zadanie 2 (3 pkt.)
# Przeanalizuj implementacje algorytmu Shora https://qiskit.org/textbook/ch-algorithms/shor.html i powtórz ją dla różnych wartości $a$.
| 11,223 |
/notes/coursera-ai-for-medicine/Course 2 - AI for Medical Prognosis/Week 1 - Diagnosing Diseases using Linear Risk Models/activities_lab/lecture_ex_03-combined_features.ipynb | 8259427b2ade836e21b0e5b96495ac5bea23994c | [] | no_license | philipsales/techinical_notes | https://github.com/philipsales/techinical_notes | 1 | 0 | null | null | null | null | Jupyter Notebook | false | false | .py | 35,136 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Course 2 week 1 lecture notebook Exercise 03
#
# <a name="combine-features"></a>
# ## Combine features
#
# In this exercise, you will practice how to combine features in a pandas dataframe. This will help you in the graded assignment at the end of the week.
#
# In addition, you will explore why it makes more sense to multiply two features rather than add them in order to create interaction terms.
#
# First, you will generate some data to work with.
# +
# Import pandas
import pandas as pd
# Import a pre-defined function that generates data
from utils import load_data
# -
# Generate features and labels
X, y = load_data(100)
X.head()
feature_names = X.columns
feature_names
# ### Combine strings
# Even though you can visually see feature names and type the name of the combined feature, you can programmatically create interaction features so that you can apply this to any dataframe.
#
# Use f-strings to combine two strings. There are other ways to do this, but Python's f-strings are quite useful.
# +
name1 = feature_names[0]
name2 = feature_names[1]
print(f"name1: {name1}")
print(f"name2: {name2}")
# -
# Combine the names of two features into a single string, separated by '_&_' for clarity
combined_names = f"{name1}_&_{name2}"
combined_names
# ### Add two columns
# - Add the values from two columns and put them into a new column.
# - You'll do something similar in this week's assignment.
X[combined_names] = X['Age'] + X['Systolic_BP']
X.head(2)
# ### Why we multiply two features instead of adding
#
# Why do you think it makes more sense to multiply two features together rather than adding them together?
#
# Please take a look at two features, and compare what you get when you add them, versus when you multiply them together.
# +
# Generate a small dataset with two features
df = pd.DataFrame({'v1': [1,1,1,2,2,2,3,3,3],
'v2': [100,200,300,100,200,300,100,200,300]
})
# add the two features together
df['v1 + v2'] = df['v1'] + df['v2']
# multiply the two features together
df['v1 x v2'] = df['v1'] * df['v2']
df
# -
# It may not be immediately apparent how adding or multiplying makes a difference; either way you get unique values for each of these operations.
#
# To view the data in a more helpful way, rearrange the data (pivot it) so that:
# - feature 1 is the row index
# - feature 2 is the column name.
# - Then set the sum of the two features as the value.
#
# Display the resulting data in a heatmap.
# Import seaborn in order to use a heatmap plot
import seaborn as sns
# +
# Pivot the data so that v1 + v2 is the value
df_add = df.pivot(index='v1',
columns='v2',
values='v1 + v2'
)
print("v1 + v2\n")
display(df_add)
print()
sns.heatmap(df_add);
# -
# Notice that it doesn't seem like you can easily distinguish clearly when you vary feature 1 (which ranges from 1 to 3), since feature 2 is so much larger in magnitude (100 to 300). This is because you added the two features together.
# #### View the 'multiply' interaction
#
# Now pivot the data so that:
# - feature 1 is the row index
# - feature 2 is the column name.
# - The values are 'v1 x v2'
#
# Use a heatmap to visualize the table.
df_mult = df.pivot(index='v1',
columns='v2',
values='v1 x v2'
)
print('v1 x v2')
display(df_mult)
print()
sns.heatmap(df_mult);
# Notice how when you multiply the features, the heatmap looks more like a 'grid' shape instead of three vertical bars.
#
# This means that you are more clearly able to make a distinction as feature 1 varies from 1 to 2 to 3.
# ### Discussion
#
# When you find the interaction between two features, you ideally hope to see how varying one feature makes an impact on the interaction term. This is better achieved by multiplying the two features together rather than adding them together.
#
# Another way to think of this is that you want to separate the feature space into a "grid", which you can do by multiplying the features together.
#
# In this week's assignment, you will create interaction terms!
# ### This is the end of this practice section.
#
# Please continue on with the lecture videos!
#
# ---
| 4,520 |
/Vehicle_Accident_ml.ipynb | 3709fb84f6b912650489297ad26e5170988e09b8 | [] | no_license | shujieli2020/Vehicle-Accident-prediction | https://github.com/shujieli2020/Vehicle-Accident-prediction | 0 | 0 | null | null | null | null | Jupyter Notebook | false | false | .py | 6,316 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# +
import numpy as np
import pandas as pd
traind=pd.read_csv(r'carstrain_2.csv')
train=traind.drop(columns='Unnamed: 0')
testd=pd.read_csv(r'carstest_2.csv')
test=testd.drop(columns='Unnamed: 0')
train=train.dropna(axis=0)
train.index=range(train.shape[0])
from sklearn.preprocessing import OneHotEncoder
enc=OneHotEncoder(categories='auto').fit(train.loc[:,'State'].values.reshape(-1,1))
result=OneHotEncoder(categories='auto').fit_transform(train.loc[:,'State'].values.reshape(-1,1))
result1=result.toarray()
newtrain=pd.concat([train,pd.DataFrame(result1)],axis=1)
newtrain.columns=['is_severe', 'State', 'std.boxcox.distance', 'Side', 'Day',
'std.temp', 'std.boxcox.humidity', 'std.boxcox.pressure',
'std.boxcox.windspeed', 'std.boxcox.precipitation', 'Amenity',
'Bump', 'Crossing', 'Give_Way', 'Junction', 'No_Exit', 'Railway',
'Roundabout', 'Station', 'Stop', 'Traffic_Calming','Traffic_Signal', 'Turning_Loop','x0_AZ', 'x0_CA', 'x0_FL', 'x0_MN', 'x0_NC', 'x0_NY', 'x0_OR',
'x0_SC', 'x0_TX', 'x0_VA']
resulttest=OneHotEncoder(categories='auto').fit_transform(test.loc[:,'State'].values.reshape(-1,1))
resultt=resulttest.toarray()
newtest=pd.concat([test,pd.DataFrame(resultt)],axis=1)
newtest.columns=['is_severe', 'State', 'std.boxcox.distance', 'Side', 'Day',
'std.temp', 'std.boxcox.humidity', 'std.boxcox.pressure',
'std.boxcox.windspeed', 'std.boxcox.precipitation', 'Amenity',
'Bump', 'Crossing', 'Give_Way', 'Junction', 'No_Exit', 'Railway',
'Roundabout', 'Station', 'Stop', 'Traffic_Calming','Traffic_Signal', 'Turning_Loop','x0_AZ', 'x0_CA', 'x0_FL', 'x0_MN', 'x0_NC', 'x0_NY', 'x0_OR',
'x0_SC', 'x0_TX', 'x0_VA']
from sklearn.preprocessing import OrdinalEncoder
newtrain.iloc[:,0]=OrdinalEncoder().fit_transform(newtrain.iloc[:,0].values.reshape(-1,1))
newtest.iloc[:,0]=OrdinalEncoder().fit_transform(newtest.iloc[:,0].values.reshape(-1,1))
from sklearn.feature_selection import SelectFromModel
from sklearn.ensemble import RandomForestClassifier as RFC
y1=newtrain.iloc[:,0].values
newtrain=newtrain.drop(columns='is_severe')
y2=newtest.iloc[:,0].values
newtest=newtest.drop(columns='is_severe')
newtrain.iloc[:,1]=OrdinalEncoder().fit_transform(newtrain.iloc[:,1].values.reshape(-1,1))
newtest.iloc[:,1]=OrdinalEncoder().fit_transform(newtest.iloc[:,1].values.reshape(-1,1))
newtrain=newtrain.drop(columns='State')
newtrain.Side=np.where(newtrain.Side == 'R', 1, 0)
newtest=newtest.drop(columns='State')
newtest.Side=np.where(newtest.Side == 'R', 1, 0)
RFC_=RFC(n_estimators=100,random_state=0)
x_embedded=SelectFromModel(RFC_).fit_transform(newtrain,y1)
tenbed=newtest.iloc[:,[0,1,3,4,5,6]]
from sklearn.model_selection import cross_val_score
from sklearn.tree import DecisionTreeClassifier
import matplotlib.pyplot as plt
clf=DecisionTreeClassifier(random_state=0)
clf=clf.fit(x_embedded,y1)
RFC_=RFC_.fit(x_embedded,y1)
score_c=clf.score(tenbed,y2)
score_r=RFC_.score(tenbed,y2)
print("single tree:{}".format(score_c),
"random forest:{}".format(score_r))
clf=DecisionTreeClassifier(random_state=0)
clf=clf.fit(newtrain,y1)
RFC_=RFC_.fit(newtrain,y1)
score_c=clf.score(newtest,y2)
score_r=RFC_.score(newtest,y2)
print("single tree:{}".format(score_c),
"random forest:{}".format(score_r))
from sklearn.linear_model import LogisticRegression as LR
from sklearn.metrics import accuracy_score
lrl2=LR(penalty="l2",solver="liblinear",C=0.5,max_iter=1000)
lrl2=lrl2.fit(newtrain,y1)
lrl2.coef_
accuracy_score(lrl2.predict(newtest),y2)
y_score = RFC_.predict_proba(newtest)
y_score_lrl2 = lrl2.predict_proba(newtest)
from sklearn.metrics import roc_curve, auc
import matplotlib.pyplot as plt
#ROC
fpr, tpr, thresholds = roc_curve(y2, y_score[:, 1])
roc_auc = auc(fpr, tpr)
def drawRoc(roc_auc,fpr,tpr):
plt.subplots(figsize=(7, 5.5))
plt.plot(fpr, tpr, color='darkorange', lw=2, label='ROC curve (area = %0.3f)' % roc_auc)
plt.plot([0, 1], [0, 1], color='navy', lw=2, linestyle='--')
plt.xlim([0.0, 1.0])
plt.ylim([0.0, 1.05])
plt.xlabel('False Positive Rate')
plt.ylabel('True Positive Rate')
plt.title('ROC Curve')
plt.legend(loc="lower right")
plt.show()
drawRoc(roc_auc, fpr, tpr)
from sklearn.metrics import confusion_matrix
#Random Forest confusion matrix
conf_matrix_rf=confusion_matrix(y2, RFC_.predict(newtest))
conf_matrix_rf
#sensitivity
conf_matrix_rf[1,1]/np.sum(conf_matrix_rf[:,1])
#specificity
conf_matrix_rf[0,0]/np.sum(conf_matrix_rf[:,0])
#Logistic Regression confusion matrix
conf_matrix_lrl2=confusion_matrix(y2, lrl2.predict(newtest))
conf_matrix_lrl2
#sensitivity
conf_matrix_lrl2[1,1]/np.sum(conf_matrix_lrl2[:,1])
#specificity
conf_matrix_lrl2[0,0]/np.sum(conf_matrix_lrl2[:,0])
ejaVu Sans', fontweight='normal')
plot_scatter(axs[0], prng)
plot_image_and_patch(axs[1], prng)
plot_bar_graphs(axs[2], prng)
plot_colored_lines(axs[3])
plot_histograms(axs[4], prng)
plot_colored_circles(axs[5], prng)
# add divider
rec = Rectangle((1 + 0.025, -2), 0.05, 16,
clip_on=False, color='gray')
axs[4].add_artist(rec)
if __name__ == "__main__":
# Set up a list of all available styles, in alphabetical order but
# the `default` and `classic` ones, which will be forced resp. in
# first and second position.
# styles with leading underscores are for internal use such as testing
# and plot types gallery. These are excluded here.
style_list = ['default', 'classic'] + sorted(
style for style in plt.style.available
if style != 'classic' and not style.startswith('_'))
# Plot a demonstration figure for every available style sheet.
for style_label in style_list:
with plt.rc_context({"figure.max_open_warning": len(style_list)}):
with plt.style.context(style_label):
plot_figure(style_label=style_label)
plt.show()
| 6,189 |
/utils/tools/notebooks/r_notebook-Copy1.ipynb | 90330640c30b8a7a711c2b908262416203c167e3 | [] | no_license | Kanaderu/iotsite | https://github.com/Kanaderu/iotsite | 3 | 3 | null | 2022-04-22T22:16:35 | 2020-11-18T05:33:24 | Jupyter Notebook | Jupyter Notebook | false | false | .r | 319,166 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .r
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: R
# language: R
# name: ir
# ---
# + jupyter={"outputs_hidden": true}
csvDataset<-read.csv("FFCC79B01604Concrete.csv")
csvDataset$Units
# -
library(rjson)
jsonDataset<-fromJSDON(file="city_country_meteor.json")
jsonDataset[1]
csvDataset<-read.csv("FFCC79B01604Concrete.csv")
plot(csvDataset$Units)
df<-read.csv("http://206.189.227.139:8181/Query/Latest/1000.csv?Units=Standard&Timezone=EST5EDT&SensorID=FFACA2A71605")
plot(y=df$Units, x=df$TimeStamp)
df<-read.csv("http://206.189.227.139:8181/Query/Latest/1000.csv?Units=Standard&Timezone=EST5EDT&SensorID=FFACA2A71605")
#plot(y=df$Units, x=df$TimeStamp)
plot(y=df$Units, x=df$TimeStamp)
df<-read.csv("http://206.189.227.139:8181/Query/Latest/1000.csv?Units=Standard&Timezone=EST5EDT&SensorID=FFACA2A71605")
plot(y=df$Units, x=df$TimeStamp, col='red')
# +
df<-read.csv("http://206.189.227.139:8181/Query/Latest/1000.csv?Units=Standard&Timezone=EST5EDT&SensorID=FFD6A0B01604")
boxplot(x=df$Units)
hist(df$Units, col = "red", breaks = 20, main="Greenroof data", xlab="Temperature")
# +
df<-read.csv("http://206.189.227.139:8181/Query/Latest/1000.csv?Units=Standard&Timezone=EST5EDT&SensorID=FFD6A0B01604")
plot(table, col.table = "red", x=df$TimeStamp, y=df$Units, main="Green Roof Plot", col.main = "red",
ylab="Temperature", col.ylab = "red", type.table="l", pch = 18, cex = 2)
# +
# Create a first line
df <- read.csv("http://206.189.227.139:8181/Query/Latest/400.csv?Units=Standard&Timezone=EST5EDT&SensorID=FFACA2A71605")
x2=df$TimeStamp
y2=df$Units
df <- read.csv("http://206.189.227.139:8181/Query/Latest/400.csv?Units=Standard&Timezone=EST5EDT&SensorID=FFD6A0B01604")
x1=df$TimeStamp
y1=df$Units
plot(x2, y2, col = "blue", xlab = "Time",
ylab = "Temperature", main = "Concrete vs Green space")
# Add a second line
lines(x1, y1, pch = 18, col = "green")
# Add a legend to the plot
legend("topleft", legend=c("Concrete", "Green space"),
col=c("black", "green"), lty = 2:1, cex=0.8)
| 2,184 |
/Homework/Day_030_HW.ipynb | 14712ca4a4bef3f0433ce9cc801a339a6d9d09db | [] | no_license | WalterWengTW/2nd-ML100Days | https://github.com/WalterWengTW/2nd-ML100Days | 0 | 0 | null | null | null | null | Jupyter Notebook | false | false | .py | 17,045 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# +
from imutils.object_detection import non_max_suppression
import numpy as np
import time
import cv2
import pytesseract
net = cv2.dnn.readNet("frozen_east_text_detection.pb")
def text_detector(image):
orig = image
(H, W) = image.shape[:2]
(newW, newH) = (320, 320)
rW = W / float(newW)
rH = H / float(newH)
image = cv2.resize(image, (newW, newH))
(H, W) = image.shape[:2]
layerNames = [
"feature_fusion/Conv_7/Sigmoid",
"feature_fusion/concat_3"]
blob = cv2.dnn.blobFromImage(image, 1.0, (W, H),
(123.68, 116.78, 103.94), swapRB=True, crop=False)
net.setInput(blob)
(scores, geometry) = net.forward(layerNames)
(numRows, numCols) = scores.shape[2:4]
rects = []
confidences = []
for y in range(0, numRows):
scoresData = scores[0, 0, y]
xData0 = geometry[0, 0, y]
xData1 = geometry[0, 1, y]
xData2 = geometry[0, 2, y]
xData3 = geometry[0, 3, y]
anglesData = geometry[0, 4, y]
# loop over the number of columns
for x in range(0, numCols):
# if our score does not have sufficient probability, ignore it
if scoresData[x] < 0.5:
continue
# compute the offset factor as our resulting feature maps will
# be 4x smaller than the input image
(offsetX, offsetY) = (x * 4.0, y * 4.0)
# extract the rotation angle for the prediction and then
# compute the sin and cosine
angle = anglesData[x]
cos = np.cos(angle)
sin = np.sin(angle)
# use the geometry volume to derive the width and height of
# the bounding box
h = xData0[x] + xData2[x]
w = xData1[x] + xData3[x]
# compute both the starting and ending (x, y)-coordinates for
# the text prediction bounding box
endX = int(offsetX + (cos * xData1[x]) + (sin * xData2[x]))
endY = int(offsetY - (sin * xData1[x]) + (cos * xData2[x]))
startX = int(endX - w)
startY = int(endY - h)
# add the bounding box coordinates and probability score to
# our respective lists
rects.append((startX, startY, endX, endY))
confidences.append(scoresData[x])
boxes = non_max_suppression(np.array(rects), probs=confidences)
for (startX, startY, endX, endY) in boxes:
startX = int(startX * rW)
startY = int(startY * rH)
endX = int(endX * rW)
endY = int(endY * rH)
boundary = 2
text = orig[startY-boundary:endY+boundary, startX - boundary:endX + boundary]
text = cv2.cvtColor(text.astype(np.uint8), cv2.COLOR_BGR2GRAY)
textRecongized = pytesseract.image_to_string(text)
cv2.rectangle(orig, (startX, startY), (endX, endY), (0, 255, 0), 3)
orig = cv2.putText(orig, textRecongized, (endX,endY+5), cv2.FONT_HERSHEY_SIMPLEX, 1, (0,0,255), 2, cv2.LINE_AA)
return orig
image0 = cv2.imread('image0.jpg')
image1 = cv2.imread('image1.jpg')
image2 = cv2.imread('image2.jpg')
image3 = cv2.imread('image3.jpg')
image4 = cv2.imread('image4.jpg')
array = [image0,image1,image2,image3,image4]
for i in range(0,2):
for img in array:
imageO = cv2.resize(img, (640,320), interpolation = cv2.INTER_AREA)
orig = cv2.resize(img, (640,320), interpolation = cv2.INTER_AREA)
textDetected = text_detector(imageO)
cv2.imshow("Orig Image",orig)
cv2.imshow("Text Detection", textDetected)
time.sleep(2)
k = cv2.waitKey(30)
if k == 27:
break
cv2.destroyAllWindows()
# -
| 3,503 |
/week4/code/CS-E5740 Complex Networks Submission 4.ipynb | d184ea0463ac7da222cbd9fc5d4fb2f7ca2636ef | [] | no_license | adamilyas/complex-networks | https://github.com/adamilyas/complex-networks | 1 | 1 | null | null | null | null | Jupyter Notebook | false | false | .py | 238,574 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# <span style="font-size:larger;">CS-E5740 Complex Networks Adam Ilyas 725819</span>
#
#
# Percolation, error & attack tolerance, epidemic
# models
#
# **Percolation theory**
#
# “order parameter” $P$: fraction of nodes in the largest connected component (LCC)
#
# Control parameters $f$: $\frac{\text{number of active links}}{\text{number of all possible links}}$
#
#
# +
import os
import random
import networkx as nx
import numpy as np
import matplotlib.pyplot as plt
import scipy.stats
from percolation_in_er_networks import *
from error_and_attack_tolerance import *
# -
# # Percolation in Erdős-Rényi (ER) networks
#
# Erdős-Rényi networks are random networks where N nodes are randomly connected such that
# the probability that a pair of nodes is linked is p.
#
# Sparse ER graph: <br>
# - average degree $\langle k \rangle$ is some fixed small number
# - the size of the network N is very large.
# - N large, p large such that
# $p(N-1) = \langle k \rangle$ stays constant
#
# Percolation properties of ER Graphs.
#
# *percolation threshold* which is the value of $\langle k \rangle$ where the giant connected component appears
# ## 1a)
# Using the idea of branching processes and assumption
# that large and sparse ER graphs are tree-like,
#
# calculate the expected number of nodes at $d$ steps away, $n_d$ , from a randomly selected node in an ER network as a function of $\langle k \rangle$ and $d$. Using this result, justify that the giant component appears in large and sparse ER
# networks when $\langle k \rangle > 1$.
#
# Hints:
# – Remember that the degree distribution of an ER network is a Poisson distribution
# when $N \rightarrow \infty$ such that $\langle k \rangle$ is constant.
#
# Hence $\langle k \rangle = \frac{\langle k^2 \rangle}{\langle k \rangle} - 1$
# $$\begin{aligned}
# n_d & = \langle q \rangle \cdot n_{d-1}\\
# & = (\frac{\langle k^2 \rangle}{\langle k \rangle} - 1) \cdot n_{d-1}\\
# & = (\frac{\langle k \rangle^2 + \langle k \rangle}{\langle k \rangle} - 1 \cdot n_{d-1}\\
# & = (\langle k \rangle + 1 -1 )\cdot n_{d-1}\\
# & = \langle k \rangle\cdot n_{d-1}
# \\\\
# n_{d-1} & = \langle k \rangle\cdot n_{d-2}\\
# n_{d-2} & = \langle k \rangle\cdot n_{d-3}\\
# \vdots &\\\\
# n_d & = \langle k \rangle^d
# \end{aligned}
# $$
#
# We recurse $d$ times
#
# where $\langle k \rangle$ is average degree, and $d$ is the number of steps
# ## 1b)
# Verify your analytical calculations for $n_d$ using numerical simulations. Calculate the
# $n_d$ value for $d \in {0 . . . 15}$, $\langle k \rangle \in \{0.5, 1, 2\}$, and starting from enough randomly selected
# nodes to get a good estimate for the expected value. Try out two network sizes: $N = 10^4$
# and $N = 10^5$ to see how the size affects the calculations.
# +
a_dir = "./assets"
if not os.path.isdir(a_dir):
os.mkdir(a_dir)
#Solution for b)-c):
fig = ER_breadth_first_search(0.5, 10**4, 10000)
fig.savefig('./assets/er_breadthfirst_05_10k.pdf')
fig = ER_breadth_first_search(1, 10**4, 10000)
fig.savefig('./assets/er_breadthfirst_1_10k.pdf')
fig = ER_breadth_first_search(2, 10**4, 100, show_netsize=True, max_depth=15)
fig.savefig('./assets/er_breadthfirst_2_10k.pdf')
fig = ER_breadth_first_search(0.5, 10**5, 10000)
fig.savefig('./assets/er_breadthfirst_05_100k.pdf')
fig = ER_breadth_first_search(1, 10**5, 10000)
fig.savefig('./assets/er_breadthfirst_1_100k.pdf')
fig = ER_breadth_first_search(2, 10**5, 100, show_netsize=True, max_depth=15)
fig.savefig('./assets/er_breadthfirst_2_100k.pdf')
# -
# ## 1c)
#
# Explore the range at which the assumption of tree-likeness of the network is valid.
#
# This can be done, for example, by calculating the number of edges that nodes at depth $d$
# have that go back to some earlier level in addition to the single edge that connects each
# node to the level $d − 1$, and
#
# reporting the average fraction of such edges to all edges that go
# from depth $d$ to earlier levels/depths.
#
# In a perfect tree this fraction is exactly 0. Comment
# on the results, and their effect on our results to exercise b). What are the other things
# that make your analytical calculation of $n_d$ to differ from your simulation results?
# **Ans:**
#
# For average_degree = 2, for both $N = 10^4 \text{ and } 10^5$, we notice from the graph above that we will expect some loops as we go into more depths.
#
# When loops occur, the average node count will not increase as fast as we will encounter the same nodes that we counted before and not count them
#
# Thus, the simulated average node count will not increase as fast as in theory
# ## 1d)
#
# Calculate the component sizes of simulated ER networks,
#
# and use this data to (loosely) verify that the percolation threshold of ER networks is at the average degree of
# $\langle k \rangle = 1$.
#
# That is, for $\langle k \rangle < 1$ the largest connected component is small (size being measured
# as number of participating nodes), and for $\langle k \rangle > 1$ it quickly reaches the network size.
# Do this by generating ER networks of size $N = 10^4$ with different average degrees: $\langle k \rangle =
# [0.00, 0.05, \cdots , 2.45, 2.50]$. For each of the ER networks, compute the size of the largest
# component and plot it against $\langle k \rangle$
ER_percolation(10**4, 2.5, 0.05)
# ## 1e)
#
# Another, a more elegant, way to find out when the percolation transition happens
# is:
#
# to try to find the point at which the possibility for the largest component size growth
# is the largest when the control parameter (here $\langle k \rangle$ or $p$) is changed very little.
#
# Think about the situation where $\langle k \rangle$ is changed so slightly that a single link is added between
# the largest component and a randomly selected node that is not in the largest component.
#
#
# The expected change in the largest component size in this situation is some times called
# susceptibility, and it should get very large values at the percolation transition point. The
# susceptibility depends on the size distribution of all the other components, and it can be
# calculated with the following formula:
#
# $$x = \frac{\sum_s s^2 C(s) - s_max^2}{\sum_s s C(s) - s_max^2}$$
#
# where $C(s)$ is the number of components with $s$ nodes. Calculate the susceptibility x for
# each network generated in exercise d), and again plot x as a function of $\langle k \rangle$. Explain the
# shape of the curve, and its implications.
# Susceptibility is large at the point when the percolation transition happens
#
# Where making a minor change to the average degree ⟨k⟩ lead to big changes in the size of largest component (LCC) in the network
#
# **Observation**
#
# We see in the curve when $⟨k⟩ \approx 1$, we see a sudden spike in both graph (by definition, both are dependant to another). As such, percolation transition happens when average degree $⟨k⟩ \approx 1$
# # Error and attack tolerance of networks
#
# Error and attack tolerance of networks are often characterized using percolation analysis, where
# links are removed from the network according to different rules.
#
# Typically this kind of analyses
# are performed on infrastructure networks, such as power-grids or road networks. In this exercise,
# we will apply this idea to a Facebook-like web-page , and focus on the role of strong and weak
# links in the network.
#
# now to remove links (one by one) from the network in the order of
# 1. descending link weight (i.e. remove strong links first),
# 2. ascending link weight (i.e. remove weak links first),
# 3. random order
# 4. descending order of edge betweenness centrality (computed for the full network at the
# beginning).
#
# While removing the links, monitor the size of the largest component S as a function of the
# fraction of removed links $f \in [0, 1]$
# ## 2a)
#
# Visualize $S$ as a function of $f$ in all four cases in one plot. There should be clear
# differences between all four curves.
# +
network_path = './OClinks_w_undir.edg' # You may want to change the path to the edge list file
network_name = 'fb-like-network'
fig = run_link_removal(network_path, network_name)
# -
# ## 2b)
#
# For which of the four approaches is the network most and least vulnerable? In other
# words, in which case does the giant component shrink fastest / slowest? Or is this even
# simple to define?
# #### Most vulnerable
# **Descending order of edge betweenness centrality** Centrality measures how important an edge is. How important meaning that this edge acts as a link between clusters of nodes and by removing this edge, the probability of breaking up a to 2 components is hight
#
# #### Least vulnerable
# **descending link weight (i.e. remove strong links first)**
# ## 2c)
#
# When comparing the removal of links in **ascending** and **descending** order strong and
# weak links first, which ones are more important for the integrity of the network? Why do
# you think this would be the case?
# **Ans**
#
# The weaker links are more important for the integrity.
#
# Removal of a weak link leads to a higher chance of severing a connection to a hub, Thus, multiple nodes that are dependant on the link will not be able to get to the node easily
# ## 2d)
#
# How would you explain the difference between
# - random removal strategy
# - removal in descending order of edge betweenness strategy?
# **Ans**
#
# Removal in descendinng order of edge betweenness follows a predefined rule, by removing edges that are deemed more important to the network first. This method is deterministic and is not random
#
# On the other hand, random removal removes edges randomly and all edges have equal chance of getting removed.
| 9,951 |
/NLP_tamrin_3_gholamrezadar_96405037.ipynb | 0a6a13e6d750e26bbbab96efc3637cf38462ab7f | [] | no_license | Gholamrezadar/Colab-notebooks | https://github.com/Gholamrezadar/Colab-notebooks | 1 | 0 | null | null | null | null | Jupyter Notebook | false | false | .py | 25,579 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Projekt - Analiza zbioru danych na podstawie Students Performance in Exams
# Bartosz Glądała
#
# ### Wstęp
# Do analizy danych w projekcie wybrano zbiór Students Performance in Exams w oparciu o stronę https://www.kaggle.com/spscientist/students-performance-in-exams?select=StudentsPerformance.csv. Dane zawierają informacje o wynikach studentów z egzaminów w Stanach Zjednoczonych w zależności od czynników społecznych. Każdy rekord jest opisany za pomocą takich wyznaczników jak na przykład: wykształcenie rodziców, pochodzenie etniczne, płeć.
#
# W ramach zadania część wykresów została przedstawiona w sposób interaktywny, co było jednym z punktów projektu. Po najechaniu na wykres wyświetlają się dodatkowe informacje dla użytkownika.
#
#
# ### Przygotowanie zabioru danych
# Pierwszym etapem analizy zbioru będzie przedstawienie zawartych w nich informacji oraz określenie jego budowy. W ramach przygotowania będzie wykonane ewentualne czyszczenie danych, jeżeli zbiór będzie tego potrzebował.
#
# Poniżej zaimportowano wszystkie wymagane biblioteki do wykonania pełnej analizy zbioru.
# +
import numpy as np
import pandas as pd
from sklearn.preprocessing import LabelEncoder
from wordcloud import WordCloud, STOPWORDS, ImageColorGenerator
import plotly.express as px
import seaborn as sns
import matplotlib.pyplot as plt
# %matplotlib inline
# -
# Pobranie zbioru danych z pliku csv oraz zapisanie go w formie data frame za pomocą biblioteki pandas
df = pd.read_csv("StudentsPerformance.csv")
# Wyświetlenie pięć losowych rekordów ze zbioru
df.sample(5)
# Sprawdzenie rozmiaru zbioru
df.shape
# Wyświetlenie informacji dotyczących kolumn opisujących każdy rekord
df.columns
# Na podstawie powyższych informacji można stwierdzić, że zbiór posiada 1000 rekordów, a każdy rekord zawiera informacje określone w 8 kolumnach, które zostały opisane poniżej:
#
# 1. Gender - płeć
# 2. Race/ethnicity - pochodzenie etniczne
# 3. Parental level of education - poziom wykształcenia rodziców,
# 4. Lunch - rodzaj obiadu spożywany przez studenta
# 5. Test preparation course - ukończenie lub brak ukończenia kursu przygotowawczego do egzaminu
# 6. Math score - wynik z matematyki
# 7. Reading score - wynik z egzaminu części czytanej
# 8. Writing score - wynik z egzaminu części pisemnej
#
# Kolejnym krokiem jest wyświetlenie informacji ogólnych o zbiorze - informacje o brakach, jak również typ danych dla każdej kolumny. Za pomocą metody describe() wygenerowano statystki opisowe, które zawierają informację o tendencji centralnej, rozproszeniu oraz kształcie zbioru z wyłączeniem wartości nieliczbowych.
df.info()
df.describe()
# Z informacji pozyskanych za pomocą metody info() można stwierdzić, że nie istnieją braki w jakiejkolwiek kolumnie. Z tego powodu nie jest wymagane czyszczenie danych, które miałoby uzupełnić ewentualne braki. Jedynymi danymi liczbowymi w kolumnach są wyniki z egzaminów z matematyki, czytania oraz pisania.
#
# W celu potwierdzenia informacji o brakach zostanie wywołana metoda do sprawdzenia wartości zerowych w kolumnach. Dodatkowo zostaną sprawdzone wartości unikatowe w każdej z kolumn.
df.isnull().sum()
#sprawdzenie czy dane są unikalne
df.nunique()
# Kolejnym krokiem w analizie będzie rysowanie wykresów w celu zaprezentowania różnych zależności zachodzących w zbiorze danych.
# Do rysowania wykresów różnego typu wykorzystano bibliotekę plotly oraz seaborn.
# Pierwszym z prezentowanych wykresów będzie sprawdzenie stosunku liczbowego ilościowego oraz procentowego
# obu płci za pomocą wykresu typu pie.
labels = ['Female', 'Male']
data=df['gender'].value_counts()
fig = px.pie(df, values=data, names=labels)
fig.show()
# W badaniu przeważają nieznaczną różnicą kobiety nad mężczyznami. Kolejnym krokiem będzie sprawdzenie liczby osób przynależących do każdej z grupy etnicznej.
#
# Do tego celu posłużono się wykresem typu histogram.
#
fig = px.histogram(df, x="race/ethnicity")
fig.show()
# Z wykresu wynika, że najlicznieją grupą w zbiorze jest C oraz D mając znaczną przewagą nad resztą.
# Najbardziej liczebnie odstającą grupą etniczną jest grupa A.
#
# Kolejnym krokiem będzie sprawdzenie liczebnie poziomu wykształcenia rodziców.
data = df['parental level of education'].value_counts().sort_index()[::-1]
fig = px.bar(df, x=data, y=data.index, orientation='h',
labels={
"x": "",
"y": "",
},)
fig.update_layout(
title_text="Parental level of eduaction",
barmode="stack",
uniformtext=dict(mode="hide", minsize=10),
)
fig.show()
# Z wykresu słupkowego o poziomie wykształcenia rodziców wynika, że wykształcenie wyższe(magisterskie oraz licencjackie) wśród rodziców jest najmniej popularne, natomiast największą popularnością cieszy się wykształcenie średnie oraz Associate's degree.
# Associate's degree jest to wykształcenie dostępne w Stanach Zjednoczonych. Wykształcenie to ma pomóc w przygotowaniu do przyszłego zawodu. Zdobycie tego tytułu jest łatwiejsze niż zdobycie tytułu licencjata.
#
# Następnie przedstawiono poziom wykształcenia rodziców na tle poszczególnych grup etnicznych. Do tego celu posłużono się dwoma typami interakcyjnych wykresów: treemap oraz sunburst.
df['total'] = 1
fig = px.treemap(df, path=['race/ethnicity', 'parental level of education'], values='total')
fig.show()
fig = px.sunburst(df, path=['race/ethnicity', 'parental level of education'], values='total')
df = df.drop(['total'], axis = 1)
fig.show()
# Za pomocą interaktywnych wykresów z łatwością można sprawdzić liczbowo poszczególny poziom wykształcenia rodziców dla każdej z grup.
#
# Z wykresu wynika, że najwięcej osób z wykształceniem wyższym znajduje się w grupach D oraz C, a z wykształceniem średnim również w C i D. Jest to spowodowane przewagą liczebną obu grup nad resztą.
#
# Poniżej przedstawiono wykres typu histogram w celu zaprezentowania odbycia kursu przygotowawczego w poszczególnych grupach.
fig = px.histogram(df, x="race/ethnicity", color = 'test preparation course')
fig.show()
# Z wykresu można stwierdzić, że ponad połowa studentów w każdej grupie nie odbyła kursu przygotowawczego przed podejściem do egzaminu.
#
# Najlepsza sytuacja w odbyciu kursu przygotowawczego ma miejsce w grupa C oraz E. Najgorsza sytuacja dotyczy ponownie grupy A. Przy analizie należy również zwrócić uwagę na liczebność każdej z grup.
#
# W tym momencie przejdziemy do danych numerycznych, które w zbiorze określone są jako wyniki testów z matematyki, pisania oraz czytania. Do zaprezentowania wyników posłużono się wykresem rozkładu z biblioteki seaborn.
sns.displot(df['math score'], kde=True, color = 'goldenrod')
sns.displot(df['reading score'], kde=True, color = 'lightcoral')
sns.displot(df['reading score'], kde=True, color = 'darkorchid')
# W każdym z przypadków można zauważyć, że najbardziej liczna grupa studentów osiągała wyniki pomiędzy 60, a 80. Wartość prawdopodobieństwa rozkładu normalnego w przypadku matematyki jest niższa w porównaniu do egzaminu z pisania oraz czytania. Można zatem stwierdzić, że dobre wyniki z matematyki były najcięższe do osiągniecia.
#
# Kolejnym typem wykresu prezentowanym poniżej będzie pairplot, który będzie miał za zadanie zaprezentowanie różnych zależności uwzględniając wszystkie typy egzaminów oraz płeć studentów.
fig = px.scatter_matrix(df,
dimensions=["math score", "reading score", "writing score"],color="gender")
fig.show()
# Z wykresu można odczytać predyspozycje płci z poszczególnych egzaminów,
# jak również zauważyć zależność otrzymania dobrego wyniku z egzaminu.
#
# Analizując wykres można zauważyć dużą relację pomiędzy dobrym wynikiem z części pisemnej jak i czytanej.
#
# Do sprawdzenia tej hipotezy posłużono się typem wykresu heatmap.
correlation = df.corr()
fig = px.imshow(correlation,color_continuous_scale='viridis')
fig.show()
# Z wykresu wynika, że jest duża relacja pomiędzy dobrym wynikiem z pisania oraz czytania, ponieważ współczynik korelacji wynosi 0,95.
# Zależność opisująca dobry wynik z egzaminu z matematyki odnoszący się do pozostałych dwóch egzaminów jest mniejsza i wynosi około 0,80 wartości współczynnika korelacji.
#
# Następnie sprawdzono wpływ poziomu wykształcenia rodziców na otrzymane rezulaty z egzaminów.
fig = px.scatter(df, x='writing score', y='reading score', color='parental level of education', opacity=0.5)
fig.show()
# Nachodząc na wybrane kropki można sprawdzić jaki wynik osiągnął student z pisania oraz czytania ze względu na poziom wykształcenia swoich rodziców. Analizując kropki z najniższymi wynikami można zauważyć, że są to zazwyczaj studenci, których rodzice mają tylko średnie wykształcenie.
#
# Kolejnym krokiem analizy było wyciągnięcie średniej z trzech wyników egzaminu oraz dodanie do wykorzystywanego zbioru danych.
df["mean score"] = ((df['math score'] + df["reading score"] + df["writing score"]) / 3).round()
df.sample(5)
# Następnie za pomocą biblioteki sklearn zakodowano dane tekstowe na liczbowe. Operacja ma na celu możliwość przejścia do bardziej złożonych analiz opartych wyłącznie na liczbach.
lc = LabelEncoder()
df['gender'] = lc.fit_transform(df['gender'])
df['race/ethnicity'] = lc.fit_transform(df['race/ethnicity'])
df['parental level of education'] = lc.fit_transform(df['parental level of education'])
df['lunch'] = lc.fit_transform(df['lunch'])
df['test preparation course'] = lc.fit_transform(df['test preparation course'])
df.sample(5)
# Do zaprezentowania uśrednionych wyników z egzaminu na podstawie pochodzenia etnicznego posłużono się wykresem pudełkowym.
fig = px.box(df, x='race/ethnicity', y='mean score', color="gender")
fig.show()
# Z wykresu typu pudełkowego możemy odczytać dla każdej grupy etnicznej wartość minimalną oraz mayksymalną, medianę, a także pierwszy i trzeci kwartał pudełka. Dodatkowo poza wąsami czasami znajdują się kropki. Są to wartości które znacznie odbiegają od tych które zostały wzięte do skontruowania pudełka. Im dłuższe pudełko tym dane są bardziej rozproszone.
#
# Ostatnim z wykorzystanych wykresów w projekcie będzie wykres typu violin. Do zaprezentowania zależności poziomu wykształcenia rodziców do uśrednionego wyniku ze wszystkich egzaminów.
fig = px.violin(df, x='parental level of education', y='mean score',
color='gender')
fig.show()
# ### Wnioski
#
# W projekcie przeanalizowano zbiór danych dotyczący wyników z egzaminów w Stanach Zjednocznych ze względu na czynniki społeczne.
# W ramach analizy przedstawiono wykresu różnego typu, które miały pokazać zachodzące zależności. Ważnym czynnikiem wpływajacym na wyniki z egzaminów jest poziom wykształcenia rodziców oraz pochodzenie etniczne. Nie mniejszą wagę ma również odbycie kursu przygotowawczego do egzaminu.
#
# Największe zależności w otrzymanych wynikach z egzaminów zauważono przy porównaniu wyników z częsci pisemnej i czytanej. Dla wszystkich studentów najciężej było osiągnać dobre wyniki z egzaminu z matematyki. Porównując wyniki egzaminów dla obu grup - kobiet i mężczyzn, łatwo można zauważyć, że to kobiety osiągają lepsze wyniki niż mężczyźni.
#
# Do prezentacji wyników posłużono się bibliotekami plotly oraz seaborn. Do bardziej zaawansowanej analizy posłużono się biblioteką sklearn, która pozwoliła przekształcić dane tekstowe na dane numeryczne. Z ich pomocą można było narysować wykresy oparte wyłącznie na danych liczbowych.
# +
plt.subplots(figsize=(10,10))
wordcloud=WordCloud(
background_color='white',
width=550,
height=420).generate(' '.join(df))
plt.imshow(wordcloud)
plt.axis('off')
plt.savefig('graph.png')
plt.show()
# -
| 11,902 |
/resultierenden_Implemmentierung_von_annotation_example1.ipynb | ce848984ced46333cefba60167fae898a13833bd | [] | no_license | darbab2s/Data-Science-Visualisation | https://github.com/darbab2s/Data-Science-Visualisation | 1 | 3 | null | null | null | null | Jupyter Notebook | false | false | .py | 837,657 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Implementation von annotation_example1.txt
#
# +
import math
import numpy as np
import random as rd
import pandas as pd
from scipy import stats
import matplotlib.pyplot as plt
import matplotlib as mat
import holoviews as hv
import vaex as vaex
import vaex.jupyter.model as vjm
import seaborn as sns
import time
# -
#
# ## Die Daten werden hier eingelesen.
# ###### Mit Übergabe als <class 'vaex.dataframe.DataFrameArrays'> mit Ausgabe der dafür benötigten Zeit
#
# Hier wurde die Datei über Vaex importiert und beim back_value gab es eine kleine Nachbereitung im Namen, da scheinbar mit dem Namen aus der letzten Spalte es zu Komplikationen kommt und das unabhängig davon dass wir hier Vaex verwenden, dies war nämlich auch beim Importieren mit Pandas auch so.
# +
start_proc = time.process_time()
df = vaex.from_csv(
'/Users/samel/Documents/Hochschule/aktuelles Semester/03. Projekt zur Datenanalyse/Datein/annotation_example1.txt',
sep=";",
header=0
)
df.rename(df.get_column_names()[6], 'backvalue')
ende_proc = time.process_time()
print('Benötigte Systemzeit: {:5.3f}s'.format(ende_proc-start_proc),'\n\nDie Länge des Datensatzes beträgt:', len(df))
df
# -
#
# ## Hier sollen die Dateien in Kategorien aufgeteilt werden
# ##### Nebenbei werden die Dateien mit den Variablen in 'df_zahl_ohne_f' übergeben, die die Datei ohne die Werte der Kategorie 'f' besteht und verwendet werden soll.
# +
f = df[df.sign == 'f'] # f => failure (ein Problem mit einem der Geräte)
n = df[df.sign == 'n'] # n => neutral (nur Lösungsmittel, keine Substanz)
p = df[df.sign == 'p'] # p => positiv (kontrolle mit -max- Effekt, bzw. verschiede Konzentrationen -> DWK)
z = df[df.sign == 'z'] # z => zero (negative kontrolle, oder zweite kontrolle, ditto)
s = df[df.sign == 's'] # s => substanz (getestetes kleines chem. Molekül)
# +
print('Länge des gesamten Datensatzes aus annotation_example1.txt:', len(df), '\n', '\n')
print('Länge der Kategorie f:', len(f), '\n')
print('Länge der Kategorie n:', len(n), '\n')
print('Länge der Kategorie p:', len(p), '\n')
print('Länge der Kategorie z:', len(z), '\n')
print('Länge der Kategorie s:', len(s), '\n')
print("Wobei wir die Kategorie 'f' vernachläsigen werden, da es sich hierbei um Gerätefehler handelt:")
print( len(df), '-' ,len(df[df.sign == 'f']), ' =', len(s) + len(z) + len(p) + len(n))
# -
#
# Hier werden Boplots geplottet.
datei = df.to_pandas_df(['pos_id', 'barcode', 'run_id', 'test_date', 'sign', 'value', 'backvalue'])
datei
# +
f = datei[ (datei['sign'] == 'f')].index
datei.drop(f, inplace=True)
# +
sns.set_theme(style="whitegrid")
ax = sns.boxplot(x="sign", y="value", data=datei)
# +
sns.set_theme(style="whitegrid")
ax = sns.boxplot(x="sign", y="backvalue", data=datei)
# -
#
# ## Inteervalle festlegen, um zukünftig Ausreißer entfernen zu können.
#
# ### Intervalsgrenzen bestimmen
# Die Intervalle beziehen sich auf die jeweilige Kategorie (n, p, z, s).
# +
print('Die Intervalle sind auf die jeweiligen signs bezogen und dem entsprechenden Feature, jedoch haben diese nicht den selben Intervall, was bei der Modelierung später angepasst werden muss:\n')
n_stdValue = n.std('value')
n_stdBackvalue = n.std('backvalue')
n_meanValue = n.mean('value')
n_meanBackvalue = n.mean('backvalue')
n_ValueGrenzen = (n_meanValue - (n_stdValue),
n_meanValue + (n_stdValue) )
n_BackvalueGrenzen = (n_meanBackvalue - (n_stdBackvalue),
n_meanBackvalue + (n_stdBackvalue) )
print('1std-n:', '\n', 'VALUE:', n_ValueGrenzen, 'BACK_VALUE:', n_BackvalueGrenzen, '\n')
n_3_ValueGrenzen = (n_meanValue - (3 * n_stdValue),
n_meanValue + (3 * n_stdValue) )
n_3_BackvalueGrenzen = (n_meanBackvalue - (3 * n_stdBackvalue),
n_meanBackvalue + (3 * n_stdBackvalue) )
print('3std-n:', '\n', 'VALUE:', n_3_ValueGrenzen, 'BACK_VALUE:', n_3_BackvalueGrenzen, '\n')
n_5_ValueGrenzen = (n_meanValue - (5 * n_stdValue),
n_meanValue + (5 * n_stdValue) )
n_5_BackvalueGrenzen = (n_meanBackvalue - (5 * n_stdBackvalue),
n_meanBackvalue + (5 * n_stdBackvalue) )
print('5std-n:', '\n', 'VALUE:', n_5_ValueGrenzen, 'BACK_VALUE:', n_5_BackvalueGrenzen, '\n', '\n', '\n')
p_stdValue = p.std('value')
p_stdBackvalue = p.std('backvalue')
p_meanValue = p.mean('value')
p_meanBackvalue = p.mean('backvalue')
p_ValueGrenzen = (p_meanValue - (p_stdValue),
p_meanValue + (p_stdValue) )
p_BackvalueGrenzen = (p_meanBackvalue - (p_stdBackvalue),
p_meanBackvalue + (p_stdBackvalue) )
print('1std-p:', '\n', 'VALUE:', p_ValueGrenzen, 'BACK_VALUE:', p_BackvalueGrenzen, '\n')
p_3_ValueGrenzen = (p_meanValue - (3 * p_stdValue),
p_meanValue + (3 * p_stdValue) )
p_3_BackvalueGrenzen = (p_meanBackvalue - (3 * p_stdBackvalue),
p_meanBackvalue + (3 * p_stdBackvalue) )
print('3std-p:', '\n', 'VALUE:', p_3_ValueGrenzen, 'BACK_VALUE:', p_3_BackvalueGrenzen, '\n')
p_5_ValueGrenzen = (p_meanValue - (5 * p_stdValue),
p_meanValue + (5 * p_stdValue) )
p_5_BackvalueGrenzen = (p_meanBackvalue - (5 * p_stdBackvalue),
p_meanBackvalue + (5 * p_stdBackvalue) )
print('5std-p:', '\n', 'VALUE:', p_5_ValueGrenzen, 'BACK_VALUE:', p_5_BackvalueGrenzen, '\n', '\n', '\n')
z_stdValue = z.std('value')
z_stdBackvalue = z.std('backvalue')
z_meanValue = z.mean('value')
z_meanBackvalue = z.mean('backvalue')
z_ValueGrenzen = (z_meanValue - (z_stdValue),
z_meanValue + (z_stdValue) )
z_BackvalueGrenzen = (z_meanBackvalue - (z_stdBackvalue),
z_meanBackvalue + (z_stdBackvalue) )
print('1std-z:', '\n', 'VALUE:', z_ValueGrenzen, 'BACK_VALUE:', z_BackvalueGrenzen, '\n')
z_3_ValueGrenzen = (z_meanValue - (3 * z_stdValue),
z_meanValue + (3 * z_stdValue) )
z_3_BackvalueGrenzen = (z_meanBackvalue - (3 * z_stdBackvalue),
z_meanBackvalue + (3 * z_stdBackvalue) )
print('3std-z:', '\n', 'VALUE:', z_3_ValueGrenzen, 'BACK_VALUE:', z_3_BackvalueGrenzen, '\n')
z_5_ValueGrenzen = (z_meanValue - (5 * z_stdValue),
z_meanValue + (5 * z_stdValue) )
z_5_BackvalueGrenzen = (z_meanBackvalue - (5 * z_stdBackvalue),
z_meanBackvalue + (5 * z_stdBackvalue) )
print('5std-z:', '\n', 'VALUE:', z_5_ValueGrenzen, 'BACK_VALUE:', z_5_BackvalueGrenzen, '\n', '\n', '\n')
s_stdValue = s.std('value')
s_stdBackvalue = s.std('backvalue')
s_meanValue = s.mean('value')
s_meanBackvalue = s.mean('backvalue')
s_ValueGrenzen = (s_meanValue - (s_stdValue),
s_meanValue + (s_stdValue) )
s_BackvalueGrenzen = (s_meanBackvalue - (s_stdBackvalue),
s_meanBackvalue + (s_stdBackvalue) )
print('1std-s:', '\n', 'VALUE:', s_ValueGrenzen, 'BACK_VALUE:', s_BackvalueGrenzen, '\n')
s_3_ValueGrenzen = (s_meanValue - (3 * s_stdValue),
s_meanValue + (3 * s_stdValue) )
s_3_BackvalueGrenzen = ( s_meanBackvalue - (3 * s_stdBackvalue),
s_meanBackvalue + (3 * s_stdBackvalue) )
print('3std-s:', '\n', 'VALUE:', s_3_ValueGrenzen, 'BACK_VALUE:', s_3_BackvalueGrenzen, '\n')
s_5_ValueGrenzen = (s_meanValue - (5 * s_stdValue),
s_meanValue + (5 * s_stdValue) )
s_5_BackvalueGrenzen = (s_meanBackvalue - (5 * s_stdBackvalue),
s_meanBackvalue + (5 * s_stdBackvalue) )
print('5std-s:', '\n', 'VALUE:', s_5_ValueGrenzen, 'BACK_VALUE:', s_5_BackvalueGrenzen, '\n')
# -
# Entsprechend der Grenzen Bereinigen
# +
print('+-(std)')
n_cleaned = n[(n['value'] <= n_ValueGrenzen[1] )
& (n['value'] >= n_ValueGrenzen[0] )
& (n['backvalue'] <= n_BackvalueGrenzen[1] )
& (n['backvalue'] >= n_BackvalueGrenzen[0] )
]
print('n:', len(n), '=> n_cleaned:', len(n_cleaned), '\n')
p_cleaned = p[(p['value'] <= p_ValueGrenzen[1] )
& (p['value'] >= p_ValueGrenzen[0] )
& (p['backvalue'] <= p_BackvalueGrenzen[1] )
& (p['backvalue'] >= p_BackvalueGrenzen[0] )
]
print('p:', len(p), '=> p_cleaned:', len(p_cleaned), '\n')
z_cleaned = z[(z['value'] <= z_ValueGrenzen[1] )
& (z['value'] >= z_ValueGrenzen[0] )
& (z['backvalue'] <= z_BackvalueGrenzen[1] )
& (z['backvalue'] >= z_BackvalueGrenzen[0] )
]
print('z:', len(z), '=> z_cleaned:', len(z_cleaned), '\n')
s_cleaned = s[(s['value'] <= s_ValueGrenzen[1] )
& (s['value'] >= s_ValueGrenzen[0] )
& (s['backvalue'] <= s_BackvalueGrenzen[1] )
& (s['backvalue'] >= s_BackvalueGrenzen[0] )
]
print('s:', len(s), '=> s_cleaned:', len(s_cleaned), '\n\n')
df_cleaned = n_cleaned.concat(p_cleaned)
df_cleaned = df_cleaned.concat(z_cleaned)
df_cleaned = df_cleaned.concat(s_cleaned)
print('result. Gesam.:',
len(df),
'=> result. clean Gesam.:',
len(df_cleaned),
'\n\n\n'
)
print('+-(3std)')
n_3_cleaned = n[(n['value'] <= n_3_ValueGrenzen[1] )
& (n['value'] >= n_3_ValueGrenzen[0] )
& (n['backvalue'] <= n_3_BackvalueGrenzen[1] )
& (n['backvalue'] >= n_3_BackvalueGrenzen[0] )
]
print('n:', len(n), '=> n_cleaned:', len(n_3_cleaned), '\n')
p_3_cleaned = p[(p['value'] <= p_3_ValueGrenzen[1] )
& (p['value'] >= p_3_ValueGrenzen[0] )
& (p['backvalue'] <= p_3_BackvalueGrenzen[1] )
& (p['backvalue'] >= p_3_BackvalueGrenzen[0] )
]
print('p:', len(p), '=> p_cleaned:', len(p_3_cleaned), '\n')
z_3_cleaned = z[(z['value'] <= z_3_ValueGrenzen[1] )
& (z['value'] >= z_3_ValueGrenzen[0] )
& (z['backvalue'] <= z_3_BackvalueGrenzen[1] )
& (z['backvalue'] >= z_3_BackvalueGrenzen[0] )
]
print('z:', len(z), '=> z_cleaned:', len(z_3_cleaned), '\n')
s_3_cleaned = s[(s['value'] <= s_3_ValueGrenzen[1] )
& (s['value'] >= s_3_ValueGrenzen[0] )
& (s['backvalue'] <= s_3_BackvalueGrenzen[1] )
& (s['backvalue'] >= s_3_BackvalueGrenzen[0] )
]
print('s:', len(s), '=> s_cleaned:', len(s_3_cleaned), '\n\n')
df_3_cleaned = n_3_cleaned.concat(p_3_cleaned)
df_3_cleaned = df_3_cleaned.concat(z_3_cleaned)
df_3_cleaned = df_3_cleaned.concat(s_3_cleaned)
print('result. Gesam.:',
len(df),
'=> result. clean Gesam.:',
len(df_3_cleaned),
'\n\n\n'
)
print('+-(5std)')
n_5_cleaned = n[(n['value'] <= n_5_ValueGrenzen[1] )
& (n['value'] >= n_5_ValueGrenzen[0] )
& (n['backvalue'] <= n_5_BackvalueGrenzen[1] )
& (n['backvalue'] >= n_5_BackvalueGrenzen[0] )
]
print('n:', len(n), '=> n_cleaned:', len(n_5_cleaned), '\n')
p_5_cleaned = p[(p['value'] <= p_5_ValueGrenzen[1] )
& (p['value'] >= p_5_ValueGrenzen[0] )
& (p['backvalue'] <= p_5_BackvalueGrenzen[1] )
& (p['backvalue'] >= p_5_BackvalueGrenzen[0] )
]
print('p:', len(p), '=> p_cleaned:', len(p_5_cleaned), '\n')
z_5_cleaned = z[(z['value'] <= z_5_ValueGrenzen[1] )
& (z['value'] >= z_5_ValueGrenzen[0] )
& (z['backvalue'] <= z_5_BackvalueGrenzen[1] )
& (z['backvalue'] >= z_5_BackvalueGrenzen[0] )
]
print('z:', len(z), '=> z_cleaned:', len(z_5_cleaned), '\n')
s_5_cleaned = s[(s['value'] <= s_5_ValueGrenzen[1] )
& (s['value'] >= s_5_ValueGrenzen[0] )
& (s['backvalue'] <= s_5_BackvalueGrenzen[1] )
& (s['backvalue'] >= s_5_BackvalueGrenzen[0] )
]
print('s:', len(s), '=> s_cleaned:', len(s_5_cleaned), '\n\n')
df_5_cleaned = n_5_cleaned.concat(p_5_cleaned)
df_5_cleaned = df_5_cleaned.concat(z_5_cleaned)
df_5_cleaned = df_5_cleaned.concat(s_5_cleaned)
print('result. Gesam.:',
len(df),
'=> result. clean Gesam.:',
len(df_5_cleaned),
'\n'
)
# -
# Erneute Bereinigung um eine gemiensame Grenzen zu finden:
# +
df_copy = df_cleaned.copy()
df_cleaned_scaled = df_copy[(df_copy['value'] <= 500 )
& (df_copy['value'] >= 0 )
& (df_copy['backvalue'] <= 200 )
& (df_copy['backvalue'] >= 0 )
]
print('df_cleaned:', len(df_cleaned), '=> df_cleaned_scaled:', len(df_cleaned_scaled), '\n')
df_copy = df_3_cleaned.copy()
df_3_cleaned_scaled = df_copy[(df_copy['value'] <= 500 )
& (df_copy['value'] >= 0 )
& (df_copy['backvalue'] <= 200 )
& (df_copy['backvalue'] >= 0 )
]
print('df_cleaned:', len(df_3_cleaned), '=> df_cleaned_scaled:', len(df_3_cleaned_scaled), '\n')
df_copy = df_5_cleaned.copy()
df_5_cleaned_scaled = df_copy[(df_copy['value'] <= 500 )
& (df_copy['value'] >= 0 )
& (df_copy['backvalue'] <= 200 )
& (df_copy['backvalue'] >= 0 )
]
print('df_cleaned:', len(df_5_cleaned), '=> df_cleaned_scaled:', len(df_5_cleaned_scaled), '\n')
# -
#
# +
n_cleaned_scaled = df_cleaned_scaled[df_cleaned_scaled.sign == 'n']
print('n:', len(n_cleaned_scaled))
p_cleaned_scaled = df_cleaned_scaled[df_cleaned_scaled.sign == 'p']
print('p:', len(p_cleaned_scaled))
z_cleaned_scaled = df_cleaned_scaled[df_cleaned_scaled.sign == 'z']
print('z:', len(z_cleaned_scaled))
s_cleaned_scaled = df_cleaned_scaled[df_cleaned_scaled.sign == 's']
print('s:', len(s_cleaned_scaled))
# +
n_3_cleaned_scaled = df_3_cleaned_scaled[df_3_cleaned_scaled.sign == 'n']
print('n:', len(n_3_cleaned_scaled))
p_3_cleaned_scaled = df_3_cleaned_scaled[df_3_cleaned_scaled.sign == 'p']
print('p:', len(p_3_cleaned_scaled))
z_3_cleaned_scaled = df_3_cleaned_scaled[df_3_cleaned_scaled.sign == 'z']
print('z:', len(z_3_cleaned_scaled))
s_3_cleaned_scaled = df_3_cleaned_scaled[df_3_cleaned_scaled.sign == 's']
print('s:', len(s_3_cleaned_scaled))
# +
n_5_cleaned_scaled = df_5_cleaned_scaled[df_5_cleaned_scaled.sign == 'n']
print('n:', len(n_5_cleaned_scaled))
p_5_cleaned_scaled = df_5_cleaned_scaled[df_5_cleaned_scaled.sign == 'p']
print('p:', len(p_5_cleaned_scaled))
z_5_cleaned_scaled = df_5_cleaned_scaled[df_5_cleaned_scaled.sign == 'z']
print('z:', len(z_5_cleaned_scaled))
s_5_cleaned_scaled = df_5_cleaned_scaled[df_5_cleaned_scaled.sign == 's']
print('s:', len(s_5_cleaned_scaled))
# -
# ## Plots
# +
print('Die Intervalle sind auf die jeweiligen signs bezogen und dem entsprechenden Feature, jedoch haben diese nicht den selben Intervall, was bei der Modelierung später angepasst werden muss:\n')
n_our_ValueGrenzen = ( 0, 500 )
n_our_BackvalueGrenzen = ( 0, 200 )
print('n_Intervall:', '\n', 'VALUE:', n_our_ValueGrenzen, 'BACK_VALUE:', n_our_BackvalueGrenzen, '\n')
p_our_ValueGrenzen = ( 0, 500 )
p_our_BackvalueGrenzen = ( 0, 200 )
print('p_Intervall:', '\n', 'VALUE:', p_our_ValueGrenzen, 'BACK_VALUE:', p_our_BackvalueGrenzen, '\n')
z_our_ValueGrenzen = ( 0, 500 )
z_our_BackvalueGrenzen = ( 0, 200 )
print('z_Intervall:', '\n', 'VALUE:', z_our_ValueGrenzen, 'BACK_VALUE:', z_our_BackvalueGrenzen, '\n')
s_our_ValueGrenzen = ( 0, 500 )
s_our_BackvalueGrenzen = ( 0, 200 )
print('s_Intervall:', '\n', 'VALUE:', s_our_ValueGrenzen, 'BACK_VALUE:', s_our_BackvalueGrenzen, '\n')
our_n = n[(n['value'] < n_our_ValueGrenzen[1] )
& (n['value'] >= n_our_ValueGrenzen[0] )
& (n['backvalue'] < n_our_BackvalueGrenzen[1] )
& (n['backvalue'] >= n_our_BackvalueGrenzen[0] )
]
print('n:', len(n), '=> n_cleaned:', len(our_n), '\n')
our_p = p[(p['value'] < p_our_ValueGrenzen[1] )
& (p['value'] >= p_our_ValueGrenzen[0] )
& (p['backvalue'] < p_our_BackvalueGrenzen[1] )
& (p['backvalue'] >= p_our_BackvalueGrenzen[0] )
]
print('p:', len(p), '=> p_cleaned:', len(our_p), '\n')
our_z = z[(z['value'] < z_our_ValueGrenzen[1] )
& (z['value'] >= z_our_ValueGrenzen[0] )
& (z['backvalue'] < z_our_BackvalueGrenzen[1] )
& (z['backvalue'] >= z_our_BackvalueGrenzen[0] )
]
print('z:', len(z), '=> z_cleaned:', len(our_z), '\n')
our_s = s[(s['value'] < s_our_ValueGrenzen[1] )
& (s['value'] >= s_our_ValueGrenzen[0] )
& (s['backvalue'] < s_our_BackvalueGrenzen[1] )
& (s['backvalue'] >= s_our_BackvalueGrenzen[0] )
]
print('s:', len(s), '=> s_cleaned:', len(our_s), '\n\n')
our_df = our_n.concat(our_p)
#df_cleaned = df_cleaned.concat(z_cleaned)
our_df = our_df.concat(our_s)
print('result. Gesam.:',
len(df),
'=> result. clean Gesam.:',
len(our_df),
'\n\n\n'
)
# +
start_proc = time.process_time()
sns.set_theme(style="whitegrid")
# n_plot
n.plot(n.backvalue,
n.value,
limits=[[0, 200],
[0, 500]],
f='log',
title='n (Anzahl: '+ str( len(our_n) ) + ')'
)
#plt.title('n (Anzahl: '+ str( len(our_n) ) + ')')
plt.show()
# p_plot
p.plot(p.backvalue,
p.value,
limits=[[0, 200],
[0, 500]],
f='log',
title='p (Anzahl: '+ str( len(our_p) ) + ')'
)
#plt.title('p (Anzahl: '+ str( len(our_p) ) + ')')
plt.show()
# z_plot
z.plot(z.backvalue,
z.value,
limits=[[0, 200],
[0, 500]],
f='log',
title='z (Anzahl: '+ str( len(our_z) ) + ')'
)
#plt.title('z (Anzahl: '+ str( len(our_z) ) + ')')
plt.show()
# s_plot
s.plot(s.backvalue,
s.value,
limits=[[0, 200],
[0, 500]],
f='log',
title='s (Anzahl: '+ str( len(our_s) ) + ')'
)
#plt.title('s (Anzahl: '+ str( len(our_s) ) + ')')
plt.show()
ende_proc = time.process_time()
print('Benötigte Systemzeit: {:5.3f}s'.format(ende_proc-start_proc))
# +
start_proc = time.process_time()
sns.set_theme(style="whitegrid")
fig, (ax1, ax2, ax3, ax4) = plt.subplots(1, 4, figsize=(24,6))
plt.sca(ax1)
# code für 1te heatmap
n.plot(n.backvalue,
n.value,
limits=[[0, 200],
[0, 500]],
f='log'
)
plt.title('n (Anzahl: '+ str( len(our_n) ) + ')')
plt.legend('(Anzahl: '+ str( len(n) ) + ')')
plt.sca(ax2)
# code für 2te heatmap
p.plot(p.backvalue,
p.value,
limits=[[0, 200],
[0, 500]],
f='log'
)
plt.title('p (Anzahl: '+ str( len(our_p) ) + ')')
plt.legend('(Anzahl: '+ str( len(p) ) + ')')
plt.sca(ax3)
# code für 3te heatmap
z.plot(z.backvalue,
z.value,
limits=[[0, 200],
[0, 500]],
f='log'
)
plt.title('z (Anzahl: '+ str( len(our_z) ) + ')')
plt.legend('(Anzahl: '+ str( len(z) ) + ')')
plt.sca(ax4)
# code für 4te heatmap
s.plot(s.backvalue,
s.value,
limits=[[0, 200],
[0, 500]],
f='log'
)
plt.title('s (Anzahl: '+ str( len(our_s) ) + ')')
plt.legend('(Anzahl: '+ str( len(s) ) + ')')
plt.show()
ende_proc = time.process_time()
print('Benötigte Systemzeit: {:5.3f}s'.format(ende_proc-start_proc))
# +
start_proc = time.process_time()
sns.set_theme(style="whitegrid")
print('+-1*std')
fig, (ax1, ax2, ax3, ax4) = plt.subplots(1, 4, figsize=(24,6))
plt.sca(ax1)
# code für 1te heatmap
n.plot(n_cleaned_scaled.backvalue,
n_cleaned_scaled.value,
limits=[[1, 200],
[1, 500]],
f='log'
)
plt.title('n (Anzahl: '+ str( len(n_cleaned_scaled) ) + ')')
plt.vlines(n_BackvalueGrenzen[0], 0, 500, linestyles ="solid", colors ="g", linewidth=4, label="untere Grenze")
plt.hlines(n_ValueGrenzen[0], 0, 200, linestyles ="solid", colors ="g", linewidth=4)
plt.vlines(n_BackvalueGrenzen[1], 0, 500, linestyles ="solid", colors ="b", linewidth=4, label="obere Grenze")
plt.hlines(n_ValueGrenzen[1], 0, 200, linestyles ="solid", colors ="b", linewidth=4)
plt.legend()
plt.sca(ax2)
# code für 2te heatmap
p.plot(p.backvalue,
p.value,
limits=[[1, 200],
[1, 500]],
f='log'
)
plt.title('p (Anzahl: '+ str( len(p_cleaned_scaled) ) + ')')
plt.vlines(p_BackvalueGrenzen[0], 0, 500, linestyles ="solid", colors ="g", linewidth=4, label="untere Grenze")
plt.hlines(p_ValueGrenzen[0], 0, 200, linestyles ="solid", colors ="g", linewidth=4)
plt.vlines(p_BackvalueGrenzen[1], 0, 500, linestyles ="solid", colors ="b", linewidth=4, label="obere Grenze")
plt.hlines(p_ValueGrenzen[1], 0, 200, linestyles ="solid", colors ="b", linewidth=4)
plt.legend()
plt.sca(ax3)
# code für 3te heatmap
z.plot(z.backvalue,
z.value,
limits=[[1, 200],
[1, 500]],
f='log'
)
plt.title('z (Anzahl: '+ str( len(z_cleaned_scaled) ) + ')')
plt.vlines(z_BackvalueGrenzen[0], 0, 500, linestyles ="solid", colors ="g", linewidth=4, label="untere Grenze")
plt.hlines(z_ValueGrenzen[0], 0, 200, linestyles ="solid", colors ="g", linewidth=4)
plt.vlines(z_BackvalueGrenzen[1], 0, 500, linestyles ="solid", colors ="b", linewidth=4, label="obere Grenze")
plt.hlines(z_ValueGrenzen[1], 0, 200, linestyles ="solid", colors ="b", linewidth=4)
plt.legend()
plt.sca(ax4)
# code für 4te heatmap
s.plot(s.backvalue,
s.value,
limits=[[1, 200],
[1, 500]],
f='log'
)
plt.title('s (Anzahl: '+ str( len(s_cleaned_scaled) ) + ')')
plt.vlines(s_BackvalueGrenzen[0], 0, 500, linestyles ="solid", colors ="g", linewidth=4, label="untere Grenze")
plt.hlines(s_ValueGrenzen[0], 0, 200, linestyles ="solid", colors ="g", linewidth=4)
plt.vlines(s_BackvalueGrenzen[1], 0, 500, linestyles ="solid", colors ="b", linewidth=4, label="obere Grenze")
plt.hlines(s_ValueGrenzen[1], 0, 200, linestyles ="solid", colors ="b", linewidth=4)
plt.legend()
plt.show()
sns.set_theme(style="whitegrid")
print('+-3*std')
fig, (ax1, ax2, ax3, ax4) = plt.subplots(1, 4, figsize=(24,6))
plt.sca(ax1)
# code für 1te heatmap
n.plot(n_cleaned_scaled.backvalue,
n_cleaned_scaled.value,
limits=[[1, 200],
[1, 500]],
f='log'
)
plt.title('n (Anzahl: '+ str( len(n_3_cleaned_scaled) ) + ')')
plt.vlines(n_3_BackvalueGrenzen[0], 0, 500, linestyles ="solid", colors ="g", linewidth=4, label="untere Grenze")
plt.hlines(n_3_ValueGrenzen[0], 0, 200, linestyles ="solid", colors ="g", linewidth=4, label="untere Grenze")
plt.vlines(n_3_BackvalueGrenzen[1], 0, 500, linestyles ="solid", colors ="b", linewidth=4, label="obere Grenze")
plt.hlines(n_3_ValueGrenzen[1], 0, 200, linestyles ="solid", colors ="b", linewidth=4, label="obere Grenze")
plt.legend()
plt.sca(ax2)
# code für 2te heatmap
p.plot(p.backvalue,
p.value,
limits=[[1, 200],
[1, 500]],
f='log'
)
plt.title('p (Anzahl: '+ str( len(p_3_cleaned_scaled) ) + ')')
#plt.vlines(p_3_BackvalueGrenzen[0], 0, 500, linestyles ="solid", colors ="g", linewidth=4, label="untere Grenze")
#plt.hlines(p_3_ValueGrenzen[0], 0, 200, linestyles ="solid", colors ="g", linewidth=4, label="untere Grenze")
plt.vlines(p_3_BackvalueGrenzen[1], 0, 500, linestyles ="solid", colors ="b", linewidth=4, label="obere Grenze")
plt.hlines(p_3_ValueGrenzen[1], 0, 200, linestyles ="solid", colors ="b", linewidth=4, label="obere Grenze")
plt.legend()
plt.sca(ax3)
# code für 3te heatmap
z.plot(z.backvalue,
z.value,
limits=[[1, 200],
[1, 500]],
f='log'
)
plt.title('z (Anzahl: '+ str( len(z_3_cleaned_scaled) ) + ')')
#plt.vlines(z_3_BackvalueGrenzen[0], 0, 500, linestyles ="solid", colors ="g", linewidth=4, label="untere Grenze")
#plt.hlines(z_3_ValueGrenzen[0], 0, 200, linestyles ="solid", colors ="g", linewidth=4, label="untere Grenze")
plt.vlines(z_3_BackvalueGrenzen[1], 0, 500, linestyles ="solid", colors ="b", linewidth=4, label="obere Grenze")
plt.hlines(z_3_ValueGrenzen[1], 0, 200, linestyles ="solid", colors ="b", linewidth=4, label="obere Grenze")
plt.legend()
plt.sca(ax4)
# code für 4te heatmap
s.plot(s.backvalue,
s.value,
limits=[[1, 200],
[1, 500]],
f='log'
)
plt.title('s (Anzahl: '+ str( len(s_3_cleaned_scaled) ) + ')')
plt.vlines(s_3_BackvalueGrenzen[0], 0, 500, linestyles ="solid", colors ="g", linewidth=4, label="untere Grenze")
plt.hlines(s_3_ValueGrenzen[0], 0, 200, linestyles ="solid", colors ="g", linewidth=4, label="untere Grenze")
plt.vlines(s_3_BackvalueGrenzen[1], 0, 500, linestyles ="solid", colors ="b", linewidth=4, label="obere Grenze")
plt.hlines(s_3_ValueGrenzen[1], 0, 200, linestyles ="solid", colors ="b", linewidth=4, label="obere Grenze")
plt.legend()
plt.show()
sns.set_theme(style="whitegrid")
print('+-5*std')
fig, (ax1, ax2, ax3, ax4) = plt.subplots(1, 4, figsize=(24,6))
plt.sca(ax1)
# code für 1te heatmap
n.plot(n_cleaned_scaled.backvalue,
n_cleaned_scaled.value,
limits=[[1, 200],
[1, 500]],
f='log'
)
plt.title('n (Anzahl: '+ str( len(n_5_cleaned_scaled) ) + ')')
plt.vlines(n_5_BackvalueGrenzen[0], 0, 500, linestyles ="solid", colors ="g", linewidth=4, label="untere Grenze")
plt.hlines(n_5_ValueGrenzen[0], 0, 200, linestyles ="solid", colors ="g", linewidth=4, label="untere Grenze")
plt.vlines(n_5_BackvalueGrenzen[1], 0, 500, linestyles ="solid", colors ="b", linewidth=4, label="obere Grenze")
plt.hlines(n_5_ValueGrenzen[1], 0, 200, linestyles ="solid", colors ="b", linewidth=4, label="obere Grenze")
plt.legend()
plt.sca(ax2)
# code für 2te heatmap
p.plot(p.backvalue,
p.value,
limits=[[1, 200],
[1, 500]],
f='log'
)
plt.title('p (Anzahl: '+ str( len(p_5_cleaned_scaled) ) + ')')
#plt.vlines(p_5_BackvalueGrenzen[0], 0, 500, linestyles ="solid", colors ="g", linewidth=4, label="untere Grenze")
#plt.hlines(p_5_ValueGrenzen[0], 0, 200, linestyles ="solid", colors ="g", linewidth=4, label="untere Grenze")
#plt.vlines(p_5_BackvalueGrenzen[1], 0, 500, linestyles ="solid", colors ="b", linewidth=4, label="obere Grenze")
plt.hlines(p_5_ValueGrenzen[1], 0, 200, linestyles ="solid", colors ="b", linewidth=4, label="obere Grenze")
plt.legend()
plt.sca(ax3)
# code für 3te heatmap
z.plot(z.backvalue,
z.value,
limits=[[1, 200],
[1, 500]],
f='log'
)
plt.title('z (Anzahl: '+ str( len(z_5_cleaned_scaled) ) + ')')
#plt.vlines(z_5_BackvalueGrenzen[0], 0, 500, linestyles ="solid", colors ="g", linewidth=4, label="untere Grenze")
#plt.hlines(z_5_ValueGrenzen[0], 0, 200, linestyles ="solid", colors ="g", linewidth=4, label="untere Grenze")
#plt.vlines(z_5_BackvalueGrenzen[1], 0, 500, linestyles ="solid", colors ="b", linewidth=4, label="obere Grenze")
plt.hlines(z_5_ValueGrenzen[1], 0, 200, linestyles ="solid", colors ="b", linewidth=4, label="obere Grenze")
plt.legend()
plt.sca(ax4)
# code für 4te heatmap
s.plot(s.backvalue,
s.value,
limits=[[1, 200],
[1, 500]],
f='log'
)
plt.title('s (Anzahl: '+ str( len(s_5_cleaned_scaled) ) + ')')
plt.vlines(s_5_BackvalueGrenzen[0], 0, 500, linestyles ="solid", colors ="g", linewidth=4, label="untere Grenze")
plt.hlines(s_5_ValueGrenzen[0], 0, 200, linestyles ="solid", colors ="g", linewidth=4, label="untere Grenze")
plt.vlines(s_5_BackvalueGrenzen[1], 0, 500, linestyles ="solid", colors ="b", linewidth=4, label="obere Grenze")
plt.hlines(s_5_ValueGrenzen[1], 0, 200, linestyles ="solid", colors ="b", linewidth=4, label="obere Grenze")
plt.legend()
plt.show()
ende_proc = time.process_time()
print('Benötigte Systemzeit: {:5.3f}s'.format(ende_proc-start_proc))
# +
sns.set_theme(style="whitegrid")
fig, (ax1, ax2, ax3, ax4) = plt.subplots(1, 4, figsize=(24,6))
plt.sca(ax1)
# code für 1te heatmap
n_cleaned_scaled.plot(n_cleaned_scaled.backvalue,
n_cleaned_scaled.value,
what=np.log(vaex.stat.count(n_cleaned_scaled.value)),
limits=[[1, 200],
[1, 500]],
f='log',
#title='n'
)
plt.sca(ax2)
# code für 2te heatmap
p_cleaned_scaled.plot(p_cleaned_scaled.backvalue,
p_cleaned_scaled.value,
what=np.log(vaex.stat.count(p_cleaned_scaled.value)),
limits=[[1, 200],
[1, 500]],
f='log',
#title='p'
)
plt.sca(ax3)
# code für 3te heatmap
z_cleaned_scaled.plot(z_cleaned_scaled.backvalue,
z_cleaned_scaled.value,
what=np.log(vaex.stat.count(z_cleaned_scaled.value)),
limits=[[1, 200],
[1, 500]],
f='log',
#title='z'
)
plt.sca(ax4)
# code für 4te heatmap
s_cleaned_scaled.plot(s_cleaned_scaled.backvalue,
s_cleaned_scaled.value,
what=np.log(vaex.stat.count(s_cleaned_scaled.value)),
limits=[[1, 200],
[1, 500]],
f='log',
#title='s'
)
plt.show()
# -
#
# +-std
# +
start_proc = time.process_time()
fig, (ax1, ax2, ax3, ax4) = plt.subplots(1, 4, figsize=(24,6))
plt.sca(ax1)
# code für 1te heatmap
plt.scatter(n_cleaned_scaled.backvalue.values,
n_cleaned_scaled.value.values,
8,
marker='+',
color='blue',
label=str(len(n_cleaned_scaled))+' n (Lösungsmittel)',
alpha=1
)
plt.xlim(1, 200)
plt.ylim(1, 500)
plt.xscale('log')
plt.yscale('log')
plt.xlabel('backvalue')
plt.ylabel('value')
plt.legend()
plt.title('n')
plt.sca(ax2)
# code für 2te heatmap
plt.scatter(p_cleaned_scaled.backvalue.values,
p_cleaned_scaled.value.values,
8,
marker='+',
color='green',
label=str(len(p_cleaned_scaled))+' p (Positiv-Kont.)',
alpha=1
)
plt.xlim(1, 200)
plt.ylim(1, 500)
plt.xscale('log')
plt.yscale('log')
plt.xlabel('backvalue')
plt.ylabel('value')
plt.legend()
plt.title('p')
plt.sca(ax3)
# code für 3te heatmap
plt.scatter(z_cleaned_scaled.backvalue.values,
z_cleaned_scaled.value.values,
8,
marker='+',
color='cyan',
label=str(len(z_cleaned_scaled))+' z (Negativ-Kont.)',
alpha=1
)
plt.xlim(1, 200)
plt.ylim(1, 500)
plt.xscale('log')
plt.yscale('log')
plt.xlabel('backvalue')
plt.ylabel('value')
plt.legend()
plt.title('z')
plt.sca(ax4)
# code für 4te heatmap
plt.scatter(s_cleaned_scaled.backvalue.values,
s_cleaned_scaled.value.values,
8,
marker='+',
color='red',
label=str(len(s_cleaned_scaled))+' s (Substanz)',
alpha=1
)
plt.xlim(1, 200)
plt.ylim(1, 500)
plt.xscale('log')
plt.yscale('log')
plt.xlabel('backvalue')
plt.ylabel('value')
plt.legend()
plt.title('s')
plt.show()
ende_proc = time.process_time()
print('Benötigte Systemzeit: {:5.3f}s'.format(ende_proc-start_proc))
# +-3std
# +
start_proc = time.process_time()
fig, (ax1, ax2, ax3, ax4) = plt.subplots(1, 4, figsize=(24,6))
plt.sca(ax1)
# code für 1te heatmap
plt.scatter(n_3_cleaned_scaled.backvalue.values,
n_3_cleaned_scaled.value.values,
8,
marker='+',
color='blue',
label=str(len(n_3_cleaned_scaled))+' n (Lösungsmittel)',
alpha=1
)
plt.xlim(1, 200)
plt.ylim(1, 500)
plt.xscale('log')
plt.yscale('log')
plt.xlabel('backvalue')
plt.ylabel('value')
plt.legend()
plt.title('n')
plt.sca(ax2)
# code für 2te heatmap
plt.scatter(p_3_cleaned_scaled.backvalue.values,
p_3_cleaned_scaled.value.values,
8,
marker='+',
color='green',
label=str(len(p_3_cleaned_scaled))+' p (Positiv-Kont.)',
alpha=1
)
plt.xlim(1, 200)
plt.ylim(1, 500)
plt.xscale('log')
plt.yscale('log')
plt.xlabel('backvalue')
plt.ylabel('value')
plt.legend()
plt.title('p')
plt.sca(ax3)
# code für 3te heatmap
plt.scatter(z_3_cleaned_scaled.backvalue.values,
z_3_cleaned_scaled.value.values,
8,
marker='+',
color='cyan',
label=str(len(z_3_cleaned_scaled))+' z (Negativ-Kont.)',
alpha=1
)
plt.xlim(1, 200)
plt.ylim(1, 500)
plt.xscale('log')
plt.yscale('log')
plt.xlabel('backvalue')
plt.ylabel('value')
plt.legend()
plt.title('z')
plt.sca(ax4)
# code für 4te heatmap
plt.scatter(s_3_cleaned_scaled.backvalue.values,
s_3_cleaned_scaled.value.values,
8,
marker='+',
color='red',
label=str(len(s_3_cleaned_scaled))+' s (Substanz)',
alpha=1
)
plt.xlim(1, 200)
plt.ylim(1, 500)
plt.xscale('log')
plt.yscale('log')
plt.xlabel('backvalue')
plt.ylabel('value')
plt.legend()
plt.title('s')
plt.show()
ende_proc = time.process_time()
print('Benötigte Systemzeit: {:5.3f}s'.format(ende_proc-start_proc))
# +-5std
# +
start_proc = time.process_time()
fig, (ax1, ax2, ax3, ax4) = plt.subplots(1, 4, figsize=(24,6))
plt.sca(ax1)
# code für 1te heatmap
plt.scatter(n_5_cleaned_scaled.backvalue.values,
n_5_cleaned_scaled.value.values,
8,
marker='+',
color='blue',
label=str(len(n_5_cleaned_scaled))+' n (Lösungsmittel)',
alpha=1
)
plt.xlim(1, 200)
plt.ylim(1, 500)
plt.xscale('log')
plt.yscale('log')
plt.xlabel('backvalue')
plt.ylabel('value')
plt.legend()
plt.title('n')
plt.sca(ax2)
# code für 2te heatmap
plt.scatter(p_5_cleaned_scaled.backvalue.values,
p_5_cleaned_scaled.value.values,
8,
marker='+',
color='green',
label=str(len(p_5_cleaned_scaled))+' p (Positiv-Kont.)',
alpha=1
)
plt.xlim(1, 200)
plt.ylim(1, 500)
plt.xscale('log')
plt.yscale('log')
plt.xlabel('backvalue')
plt.ylabel('value')
plt.legend()
plt.title('p')
plt.sca(ax3)
# code für 3te heatmap
plt.scatter(z_5_cleaned_scaled.backvalue.values,
z_5_cleaned_scaled.value.values,
8,
marker='+',
color='cyan',
label=str(len(z_3_cleaned_scaled))+' z (Negativ-Kont.)',
alpha=1
)
plt.xlim(1, 200)
plt.ylim(1, 500)
plt.xscale('log')
plt.yscale('log')
plt.xlabel('backvalue')
plt.ylabel('value')
plt.legend()
plt.title('z')
plt.sca(ax4)
# code für 4te heatmap
plt.scatter(s_5_cleaned_scaled.backvalue.values,
s_5_cleaned_scaled.value.values,
8,
marker='+',
color='red',
label=str(len(s_5_cleaned_scaled))+' s (Substanz)',
alpha=1
)
plt.xlim(1, 200)
plt.ylim(1, 500)
plt.xscale('log')
plt.yscale('log')
plt.xlabel('backvalue')
plt.ylabel('value')
plt.legend()
plt.title('s')
plt.show()
ende_proc = time.process_time()
print('Benötigte Systemzeit: {:5.3f}s'.format(ende_proc-start_proc))
# -
#
#
# +
start_proc = time.process_time()
fig = plt.figure()
ax1 = plt.gca()
ax1.scatter(s.backvalue.values,
s.value.values,
4,
marker="+",
color='red',
label=('s', len(s)),
alpha=1
)
ax1.set_xlim(1, 200)
ax1.set_ylim(1, 500)
ax1.set_yscale('log')
ax1.set_xscale('log')
ax1.set_xlabel('backvalue')
ax1.set_ylabel('value')
ax1.set_aspect(0.5)
ax2 = plt.gca()
ax2.scatter(n.backvalue.values,
n.value.values,
4,
marker="+",
color='blue',
label=('n', len(n)),
alpha=1
)
ax2.set_xlim(1, 200)
ax2.set_ylim(1, 500)
ax2.set_yscale('log')
ax2.set_xscale('log')
ax2.set_xlabel('backvalue')
ax2.set_ylabel('value')
ax2.set_aspect(0.5)
ax3 = plt.gca()
ax3.scatter(p.backvalue.values,
p.value.values,
4,
marker="+",
color='green',
label=('p', len(p)),
alpha=1
)
ax3.set_xlim(1, 200)
ax3.set_ylim(1, 500)
ax3.set_yscale('log')
ax3.set_xscale('log')
ax3.set_xlabel('backvalue')
ax3.set_ylabel('value')
ax3.set_aspect(0.5)
ax4 = plt.gca()
ax4.scatter(z.backvalue.values,
z.value.values,
4,
marker="+",
color='cyan',
label=('z', len(z)),
alpha=1
)
ax4.set_xlim(1, 200)
ax4.set_ylim(1, 500)
ax4.set_yscale('log')
ax4.set_xscale('log')
ax4.set_xlabel('backvalue')
ax4.set_ylabel('value')
ax4.set_aspect(0.5)
leg = plt.legend(loc='best', bbox_to_anchor=(1.05, 1), shadow=True)
leg.get_frame().set_alpha(0.5)
plt.show()
ende_proc = time.process_time()
print('Benötigte Systemzeit: {:5.3f}s'.format(ende_proc-start_proc))
# -
# %matplotlib inline
import numpy as np
from fcmeans import FCM
from matplotlib import pyplot as plt
df_copy = df_cleaned_scaled.to_copy(column_names=('value', 'backvalue'))
df_numpy = df_copy.values
# +
start_proc = time.process_time()
fcm = FCM(n_clusters=4)
fcm.fit(df_numpy)
ende_proc = time.process_time()
print('Benötigte Systemzeit: {:5.3f}s'.format(ende_proc-start_proc))
# +
start_proc = time.process_time()
# outputs
fcm_centers = fcm.centers
fcm_labels = fcm.predict(df_numpy)
# plot result
#f, axes = plt.subplots(1, 2, figsize=(11,5))
#axes[0].scatter(X[:,0], X[:,1], alpha=.1)
plt.scatter(df_numpy[:,0], df_numpy[:,1], c=fcm_labels, alpha=.1)
plt.scatter(fcm_centers[:,0], fcm_centers[:,1], marker="+", s=500, c='w')
#plt.savefig('images/basic-clustering-output.jpg')
plt.show()
ende_proc = time.process_time()
print('Benötigte Systemzeit: {:5.3f}s'.format(ende_proc-start_proc))
# -
| 38,320 |
/Module6/NeuralNetwoks.ipynb | 10813e6dae1378e304b655a5757e8d202c2fd113 | [] | no_license | sriramsoftware/Principles-of-Machine-Learning-Python | https://github.com/sriramsoftware/Principles-of-Machine-Learning-Python | 0 | 0 | null | null | null | null | Jupyter Notebook | false | false | .py | 232,701 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Python 3.5
# language: python
# name: python3
# ---
# + [markdown] deletable=true editable=true
# # Classification with Neural Networks
#
# **Neural networks** are a powerful set of machine learning algorithms. Neural network use one or more **hidden layers** of multiple **hidden units** to perform **function approximation**. The use of multiple hidden units in one or more layers, allows neural networks to approximate complex functions. Neural network models capable of approximating complex functions are said to have high **model capacity**. This property allows neural networks to solve complex machine learning problems.
#
# However, because of the large number of hidden units, neural networks have many **weights** or **parameters**. This situation often leads to **over-fitting** of neural network models, which limits generalization. Thus, finding optimal hyperparameters when fitting neural network models is essential for good performance.
#
# An additional issue with neural networks is **computational complexity**. Many optimization iterations are required. Each optimization iteration requires the update of a large number of parameters.
# + [markdown] deletable=true editable=true
# ## Example: Iris dataset
#
# As a first example you will use neutral network models to classify the species of iris flowers using the famous iris dataset.
#
# As a first step, execute the code in the cell below to load the required packages to run the rest of this notebook.
# + deletable=true editable=true
from sklearn.neural_network import MLPClassifier
from sklearn import preprocessing
from statsmodels.api import datasets
import sklearn.model_selection as ms
import sklearn.metrics as sklm
import matplotlib.pyplot as plt
import pandas as pd
import numpy as np
import numpy.random as nr
# %matplotlib inline
# + [markdown] deletable=true editable=true
# To get a feel for these data, you will now load and plot them. The code in the cell below does the following:
#
# 1. Loads the iris data as a Pandas data frame.
# 2. Adds column names to the data frame.
# 3. Displays all 4 possible scatter plot views of the data.
#
# Execute this code and examine the results.
# + deletable=true editable=true
def plot_iris(iris):
'''Function to plot iris data by type'''
setosa = iris[iris['Species'] == 'setosa']
versicolor = iris[iris['Species'] == 'versicolor']
virginica = iris[iris['Species'] == 'virginica']
fig, ax = plt.subplots(2, 2, figsize=(12,12))
x_ax = ['Sepal_Length', 'Sepal_Width']
y_ax = ['Petal_Length', 'Petal_Width']
for i in range(2):
for j in range(2):
ax[i,j].scatter(setosa[x_ax[i]], setosa[y_ax[j]], marker = 'x')
ax[i,j].scatter(versicolor[x_ax[i]], versicolor[y_ax[j]], marker = 'o')
ax[i,j].scatter(virginica[x_ax[i]], virginica[y_ax[j]], marker = '+')
ax[i,j].set_xlabel(x_ax[i])
ax[i,j].set_ylabel(y_ax[j])
## Import the dataset
iris = datasets.get_rdataset("iris")
iris.data.columns = ['Sepal_Length', 'Sepal_Width', 'Petal_Length', 'Petal_Width', 'Species']
## Plot views of the iris data
plot_iris(iris.data)
# + [markdown] deletable=true editable=true
# You can see that Setosa (in blue) is well separated from the other two categories. The Versicolor (in orange) and the Virginica (in green) show considerable overlap. The question is how well our classifier will seperate these categories.
#
# Scikit Learn classifiers require numerically coded numpy arrays for the features and as a label. The code in the cell below does the following processing:
# 1. Creates a numpy array of the features.
# 2. Numerically codes the label using a dictionary lookup, and converts it to a numpy array.
#
# Execute this code.
# + deletable=true editable=true
Features = np.array(iris.data[['Sepal_Length', 'Sepal_Width', 'Petal_Length', 'Petal_Width']])
levels = {'setosa':0, 'versicolor':1, 'virginica':2}
Labels = np.array([levels[x] for x in iris.data['Species']])
# + [markdown] deletable=true editable=true
# Next, execute the code in the cell below to split the dataset into test and training set. Notice that unusually, 100 of the 150 cases are being used as the test dataset.
# + deletable=true editable=true
## Randomly sample cases to create independent training and test data
nr.seed(1115)
indx = range(Features.shape[0])
indx = ms.train_test_split(indx, test_size = 100)
X_train = Features[indx[0],:]
y_train = np.ravel(Labels[indx[0]])
X_test = Features[indx[1],:]
y_test = np.ravel(Labels[indx[1]])
# + [markdown] deletable=true editable=true
# As is always the case with machine learning, numeric features must be scaled. The code in the cell below performs the following processing:
#
# 1. A Zscore scale object is defined using the `StandarScaler` function from the Scikit Learn preprocessing package.
# 2. The scaler is fit to the training features. Subsequently, this scaler is used to apply the same scaling to the test data and in production.
# 3. The training features are scaled using the `transform` method.
#
# Execute this code.
# + deletable=true editable=true
scale = preprocessing.StandardScaler()
scale.fit(X_train)
X_train = scale.transform(X_train)
# + [markdown] deletable=true editable=true
# Now you will define and fit a neural network model. The code in the cell below defines a single hidden layer neural network model with 50 units. The code uses the MLPClassifer function from the Scikit Lean neural_network package. The model is then fit. Execute this code.
# + deletable=true editable=true
nr.seed(1115)
nn_mod = MLPClassifier(hidden_layer_sizes = (50,))
nn_mod.fit(X_train, y_train)
# + [markdown] deletable=true editable=true
# Notice that the many neural network model object hyperparameters are displayed. Optimizing these parameters for a given situation can be quite time consuming.
#
# Next, the code in the cell below performs the following processing to score the test data subset:
# 1. The test features are scaled using the scaler computed for the training features.
# 2. The `predict` method is used to compute the scores from the scaled features.
#
# Execute this code.
# + deletable=true editable=true
X_test = scale.transform(X_test)
scores = nn_mod.predict(X_test)
# + [markdown] deletable=true editable=true
# It is time to evaluate the model results. Keep in mind that the problem has been made difficult deliberately, by having more test cases than training cases.
#
# The iris data has three species categories. Therefore it is necessary to use evaluation code for a three category problem. The function in the cell below extends code from pervious labs to deal with a three category problem. Execute this code and examine the results.
# + deletable=true editable=true
def print_metrics_3(labels, scores):
conf = sklm.confusion_matrix(labels, scores)
print(' Confusion matrix')
print(' Score Setosa Score Versicolor Score Virginica')
print('Actual Setosa %6d' % conf[0,0] + ' %5d' % conf[0,1] + ' %5d' % conf[0,2])
print('Actual Versicolor %6d' % conf[1,0] + ' %5d' % conf[1,1] + ' %5d' % conf[1,2])
print('Actual Vriginica %6d' % conf[2,0] + ' %5d' % conf[2,1] + ' %5d' % conf[2,2])
## Now compute and display the accuracy and metrics
print('')
print('Accuracy %0.2f' % sklm.accuracy_score(labels, scores))
metrics = sklm.precision_recall_fscore_support(labels, scores)
print(' ')
print(' Setosa Versicolor Virginica')
print('Num case %0.2f' % metrics[3][0] + ' %0.2f' % metrics[3][1] + ' %0.2f' % metrics[3][2])
print('Precision %0.2f' % metrics[0][0] + ' %0.2f' % metrics[0][1] + ' %0.2f' % metrics[0][2])
print('Recall %0.2f' % metrics[1][0] + ' %0.2f' % metrics[1][1] + ' %0.2f' % metrics[1][2])
print('F1 %0.2f' % metrics[2][0] + ' %0.2f' % metrics[2][1] + ' %0.2f' % metrics[2][2])
print_metrics_3(y_test, scores)
# + [markdown] deletable=true editable=true
# Examine these results. Notice the following:
# 1. The confusion matrix has dimension 3X3. You can see that most cases are correctly classified.
# 2. The overall accuracy is 0.88. Since the classes are roughly balanced, this metric indicates relatively good performance of the classifier, particularly since it was only trained on 50 cases.
# 3. The precision, recall and F1 for each of the classes is relatively good. Versicolor has the worst metrics since it has the largest number of misclassified cases.
#
# To get a better feel for what the classifier is doing, the code in the cell below displays a set of plots showing correctly (as '+') and incorrectly (as 'o') cases, with the species color-coded. Execute this code and examine the results.
# + deletable=true editable=true
def plot_iris_score(iris, y_test, scores):
'''Function to plot iris data by type'''
## Find correctly and incorrectly classified cases
true = np.equal(scores, y_test).astype(int)
## Create data frame from the test data
iris = pd.DataFrame(iris)
levels = {0:'setosa', 1:'versicolor', 2:'virginica'}
iris['Species'] = [levels[x] for x in y_test]
iris.columns = ['Sepal_Length', 'Sepal_Width', 'Petal_Length', 'Petal_Width', 'Species']
## Set up for the plot
fig, ax = plt.subplots(2, 2, figsize=(12,12))
markers = ['o', '+']
x_ax = ['Sepal_Length', 'Sepal_Width']
y_ax = ['Petal_Length', 'Petal_Width']
for t in range(2): # loop over correct and incorect classifications
setosa = iris[(iris['Species'] == 'setosa') & (true == t)]
versicolor = iris[(iris['Species'] == 'versicolor') & (true == t)]
virginica = iris[(iris['Species'] == 'virginica') & (true == t)]
# loop over all the dimensions
for i in range(2):
for j in range(2):
ax[i,j].scatter(setosa[x_ax[i]], setosa[y_ax[j]], marker = markers[t], color = 'blue')
ax[i,j].scatter(versicolor[x_ax[i]], versicolor[y_ax[j]], marker = markers[t], color = 'orange')
ax[i,j].scatter(virginica[x_ax[i]], virginica[y_ax[j]], marker = markers[t], color = 'green')
ax[i,j].set_xlabel(x_ax[i])
ax[i,j].set_ylabel(y_ax[j])
plot_iris_score(X_test, y_test, scores)
# + [markdown] deletable=true editable=true
# Examine these plots. You can see how the classifier has divided the feature space between the classes. Notice that most of the errors occur in the overlap region between Virginica and Versicolor. This behavior is to be expected. There is an error in classifying Setosa which is a bit surprising, and which probably arises from the projection of the division between classes.
# + [markdown] deletable=true editable=true
# Is it possible that a more complex neural network would separate these cases better? The more complex model should have greater model capacity, but will be more susceptible to over-fitting. The code in the cell below uses a neural network with 2 hidden layers and 100 units per layer, coded as (100,100). This model is fit with the training data and displays the evaluation of the model.
#
# Execute this code, and answer **Question 1** on the course page.
# + deletable=true editable=true
nr.seed(1115)
nn_mod = MLPClassifier(hidden_layer_sizes = (100,100),
max_iter=300)
nn_mod.fit(X_train, y_train)
scores = nn_mod.predict(X_test)
print_metrics_3(y_test, scores)
plot_iris_score(X_test, y_test, scores)
# + [markdown] deletable=true editable=true
# These are remarkably good results. Apparently, adding additional model capacity allowed the neural network model to perform exceptionally well. There are only 7 misclassified cases, giving an overall accuracy of 0.93.
# + [markdown] deletable=true editable=true
# ## Another example
#
# Now, you will try a more complex example using the credit scoring data. You will use the prepared data which had the following preprocessing:
# 1. Cleaning missing values.
# 2. Aggregating categories of certain categorical variables.
# 3. Encoding categorical variables as binary dummy variables.
# 4. Standardizing numeric variables.
#
# Execute the code in the cell below to load the features and labels as numpy arrays for the example.
# + deletable=true editable=true
Features = np.array(pd.read_csv('Credit_Features.csv'))
Labels = np.array(pd.read_csv('Credit_Labels.csv'))
Labels = Labels.reshape(Labels.shape[0],)
print(Features.shape)
print(Labels.shape)
# + [markdown] deletable=true editable=true
# Neural network training is known to be problematic when there is significant class imbalance. Unfortunately, neural networks have no method for weighting cases. Some alternatives are:
# 1. **Impute** new values using a statistical algorithm.
# 2. **Undersample** the majority cases. For this method a number of the cases equal to the minority case are Bernoulli sampled from the majority case.
# 3. **Oversample** the minority cases. For this method the number of minority cases are resampled until they equal the number of majority cases.
#
# The code in the cell below oversamples the minority cases; bad credit customers. Execute this code to create a data set with balanced cases.
# + deletable=true editable=true
temp_Labels = Labels[Labels == 1]
temp_Features = Features[Labels == 1,:]
temp_Features = np.concatenate((Features, temp_Features), axis = 0)
temp_Labels = np.concatenate((Labels, temp_Labels), axis = 0)
print(temp_Features.shape)
print(temp_Labels.shape)
# + [markdown] deletable=true editable=true
# Nested cross validation is used to estimate the optimal hyperparameters and perform model selection for a neural network model. 3 fold cross validation is used since training neural networks is computationally intensive. Additional folds would give better estimates but at the cost of greater computation time. Execute the code in the cell below to define inside and outside fold objects.
# + deletable=true editable=true
nr.seed(123)
inside = ms.KFold(n_splits=3, shuffle = True)
nr.seed(321)
outside = ms.KFold(n_splits=3, shuffle = True)
# + [markdown] deletable=true editable=true
# The code in the cell below estimates the best hyperparameters using 3 fold cross validation. In the interest of computational efficiency, values for only 4 parameters will be searched. There are several points to note here:
# 1. In this case, a grid of four hyperparameters:
# - **alpha** is the l2 regularization hyperparameter,
# - **early_stopping** determines when the training metric becomes worse following an iteration of the optimization algorithm stops the training at the previous iteration. Early stopping is a powerful method to prevent over-fitting of machine learning models in general and neural networks in particular,
# - **beta_1** and **beta_2** are hyperparameters that control the adaptive learning rate used by the **Adam** optimizer,
# 3. The model is fit on the grid, and
# 4. The best estimated hyperparameters are printed.
#
# This code searches over a 3X3X3X2 or 54 element grid using 3 fold cross validation. Using even this modest search grid and number of folds requires the model to be trained 162 times. Execute this code and examine the result, but expect execution to take some time.
#
# Once you have executed the code, answer **Question 2** on the course page.
# + deletable=true editable=true
## Define the dictionary for the grid search and the model object to search on
param_grid = {#"alpha":[0.0000001,0.000001,0.00001],
#"early_stopping":[True, False],
"beta_1":[0.95,0.90,0.80],
"beta_2":[0.999,0.9,0.8]}
## Define the Neural Network model
nn_clf = MLPClassifier(hidden_layer_sizes = (100,100),
max_iter=300)
## Perform the grid search over the parameters
nr.seed(3456)
nn_clf = ms.GridSearchCV(estimator = nn_clf, param_grid = param_grid,
cv = inside, # Use the inside folds
scoring = 'recall',
return_train_score = True)
nr.seed(6677)
nn_clf.fit(temp_Features, temp_Labels)
#print(nn_clf.best_estimator_.alpha)
#print(nn_clf.best_estimator_.early_stopping)
print(nn_clf.best_estimator_.beta_1)
print(nn_clf.best_estimator_.beta_2)
# + [markdown] deletable=true editable=true
# Now, you will run the code in the cell below to perform the outer cross validation of the model. The multiple trainings of this model will take some time.
# + deletable=true editable=true
nr.seed(498)
cv_estimate = ms.cross_val_score(nn_clf, temp_Features, temp_Labels,
cv = outside) # Use the outside folds
print('Mean performance metric = %4.3f' % np.mean(cv_estimate))
print('SDT of the metric = %4.3f' % np.std(cv_estimate))
print('Outcomes by cv fold')
for i, x in enumerate(cv_estimate):
print('Fold %2d %4.3f' % (i+1, x))
# + [markdown] deletable=true editable=true
# Examine these results. Notice that the standard deviation of the mean of Recall is an order of magnitude less than the mean itself. This indicates that this model is likely to generalize well, but the level of performance is still unclear.
#
# Now, you will build and test a model using the estimated optimal hyperparameters. However, there is a complication. The training data subset must have the minority case oversampled. Execute the code in the cell below to create training and testing dataset, with oversampled minority cases for the training subset.
# + deletable=true editable=true
## Randomly sample cases to create independent training and test data
nr.seed(1115)
indx = range(Features.shape[0])
indx = ms.train_test_split(indx, test_size = 300)
X_train = Features[indx[0],:]
y_train = np.ravel(Labels[indx[0]])
X_test = Features[indx[1],:]
y_test = np.ravel(Labels[indx[1]])
## Oversample the minority case for the training data
y_temp = y_train[y_train == 1]
X_temp = X_train[y_train == 1,:]
X_train = np.concatenate((X_train, X_temp), axis = 0)
y_train = np.concatenate((y_train, y_temp), axis = 0)
# + [markdown] deletable=true editable=true
# The code in the cell below defines a neural network model object using the estimated optimal model hyperparameters and then fits the model to the training data. Execute this code.
# + deletable=true editable=true
nr.seed(1115)
nn_mod = MLPClassifier(hidden_layer_sizes = (100,100),
#alpha = nn_clf.best_estimator_.alpha,
#early_stopping = nn_clf.best_estimator_.early_stopping,
beta_1 = nn_clf.best_estimator_.beta_1,
beta_2 = nn_clf.best_estimator_.beta_2,
max_iter = 300)
nn_mod.fit(X_train, y_train)
# + [markdown] deletable=true editable=true
# As expected, the hyperparameters of the neural network model object reflect those specified.
#
# The code in the cell below scores and prints evaluation metrics for the model, using the test data subset.
#
# Execute this code, examine the results, and answer **Question 3** on the course page.
# + deletable=true editable=true
def score_model(probs, threshold):
return np.array([1 if x > threshold else 0 for x in probs[:,1]])
def print_metrics(labels, probs, threshold):
scores = score_model(probs, threshold)
metrics = sklm.precision_recall_fscore_support(labels, scores)
conf = sklm.confusion_matrix(labels, scores)
print(' Confusion matrix')
print(' Score positive Score negative')
print('Actual positive %6d' % conf[0,0] + ' %5d' % conf[0,1])
print('Actual negative %6d' % conf[1,0] + ' %5d' % conf[1,1])
print('')
print('Accuracy %0.2f' % sklm.accuracy_score(labels, scores))
print('AUC %0.2f' % sklm.roc_auc_score(labels, probs[:,1]))
print('Macro precision %0.2f' % float((float(metrics[0][0]) + float(metrics[0][1]))/2.0))
print('Macro recall %0.2f' % float((float(metrics[1][0]) + float(metrics[1][1]))/2.0))
print(' ')
print(' Positive Negative')
print('Num case %6d' % metrics[3][0] + ' %6d' % metrics[3][1])
print('Precision %6.2f' % metrics[0][0] + ' %6.2f' % metrics[0][1])
print('Recall %6.2f' % metrics[1][0] + ' %6.2f' % metrics[1][1])
print('F1 %6.2f' % metrics[2][0] + ' %6.2f' % metrics[2][1])
probabilities = nn_mod.predict_proba(X_test)
print_metrics(y_test, probabilities, 0.5)
# + [markdown] deletable=true editable=true
# The performance of the neural network model is less than ideal. For the negative (bad credit) case the recall is perhaps adequate, but the precision is poor. Perhaps the oversampling does not help much in this case. Challenge yourself - try and perform the undersampling method and compare the result!
# + [markdown] deletable=true editable=true
# ## Summary
#
# In this lab you have accomplished the following:
# 1. Used neural models to classify the cases of the iris data. The model with greater capacity achieved significantly better results.
# 2. Used 3 fold to find estimated optimal hyperparameters for a neural network model to classify credit risk cases. Oversampling of the minority case for the training data was required to deal with the class imbalance. Despite this approach, the results achieved are marginal at best. Perhaps, a model with greater capacity would achieve better results, or a different approach to dealing with class imbalance would be more successful.
# + deletable=true editable=true
| 22,016 |
/PruebaBavaria.ipynb | b78ee6e3908fa9eb60fa20346fa328520ec94d60 | [] | no_license | mpachonm/Bavaria_Prueba | https://github.com/mpachonm/Bavaria_Prueba | 0 | 0 | null | null | null | null | Jupyter Notebook | false | false | .py | 281,988 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# #!pip install nltk
# #!pip install emoji
# #!pip install xgboost
from wordcloud import WordCloud, STOPWORDS
import requests
import pandas as pd
import nltk
import seaborn as sns
import matplotlib.pyplot as plt
sns.set(style="darkgrid")
sns.set(font_scale=1.3)
import numpy as np
pd.set_option('display.max_colwidth', None)
from time import time
import re
import string
import os
import emoji
from sklearn.feature_extraction.text import TfidfVectorizer
from sklearn.model_selection import train_test_split,cross_val_score
from sklearn import svm
from sklearn.metrics import accuracy_score, precision_score, recall_score, f1_score,confusion_matrix,classification_report
from sklearn.tree import DecisionTreeClassifier
from sklearn.ensemble import RandomForestClassifier
from sklearn.base import BaseEstimator,TransformerMixin
from nltk.corpus import stopwords
from nltk.stem import PorterStemmer
from nltk.tokenize import word_tokenize
import warnings
warnings.filterwarnings('ignore')
np.random.seed(37)
# +
#result=requests.get('http://159.65.217.53:3001/brandbuzz/segusimiento/Pruebatecnica/LWL05x51mHOMT9GtOqnHHgqWzc72')
# -
#df = pd.DataFrame(result.json())
#df.to_csv('datos_bavaria.csv', sep='\t', encoding='utf-8')
df=pd.read_csv('C:/Users/manuel.pachon/Desktop/datos_bavaria_1.csv', encoding='latin-1')
df[['Campañas','SENTIMENT','CONTENT','MEDIA_PROVIDER']].head()
# # **Análisis descriptivo de los datos**
# Se observa que hay de 563 reacciones positivas, 374 neutrales y 63 negativas.
print(df[['SENTIMENT']].value_counts())
sent=sns.catplot(x="SENTIMENT", data=df, kind="count", height=6, aspect=1.5, palette="PuBuGn_d")
sent.fig.subplots_adjust(top=0.9) # adjust the Figure in rp
sent.fig.suptitle('Distribución de los sentimientos')
sent.set_axis_labels("", "Total")
plt.show();
camp=sns.catplot(x="Campañas", data=df, kind="count", height=6, aspect=1.5, palette="PuBuGn_d")
camp.fig.subplots_adjust(top=0.9) # adjust the Figure in rp
camp.fig.suptitle('Distribución de las campañas')
camp.set_axis_labels("", "Total")
camp.set_xticklabels(rotation=90)
med=sns.catplot(x="MEDIA_PROVIDER", data=df, kind="count", height=6, aspect=1.5, palette="PuBuGn_d")
med.fig.subplots_adjust(top=0.9) # adjust the Figure in rp
med.fig.suptitle('Distribución de MEDIA_PROVIDER')
med.set_axis_labels("", "Total")
med.set_xticklabels(rotation=90)
# Casi todos los comentarios de CONTENT pertenecen a la campaña USUARIO y al MEDIA_PROVIDER facebook.
# # Diferencias entre las diferentes categorías de SENTIMENT
class TextCounts(BaseEstimator, TransformerMixin):
def count_regex(self, pattern, tweet):
return len(re.findall(pattern, tweet))
def fit(self, X, y=None, **fit_params):
# fit method is used when specific operations need to be done on the train data, but not on the test data
return self
def transform(self, X, **transform_params):
count_words = X.apply(lambda x: self.count_regex(r'\w+', x))
count_mentions = X.apply(lambda x: self.count_regex(r'@\w+', x))
count_hashtags = X.apply(lambda x: self.count_regex(r'#\w+', x))
count_capital_words = X.apply(lambda x: self.count_regex(r'\b[A-Z]{2,}\b', x))
count_excl_quest_marks = X.apply(lambda x: self.count_regex(r'!|\?', x))
count_urls = X.apply(lambda x: self.count_regex(r'http.?://[^\s]+[\s]?', x))
# We will replace the emoji symbols with a description, which makes using a regex for counting easier
# Moreover, it will result in having more words in the tweet
count_emojis = X.apply(lambda x: emoji.demojize(x)).apply(lambda x: self.count_regex(r':[a-z_&]+:', x))
df = pd.DataFrame({'conteo_palabras': count_words
, 'conteo_menciones': count_mentions
, 'conteo_hashtags': count_hashtags
, 'conteo_palabras_mayusculas': count_capital_words
, 'conteo_exclam_interog': count_excl_quest_marks
, 'conteo_urls': count_urls
, 'conteo_emojis': count_emojis
})
return df
tc = TextCounts()
df_eda = tc.fit_transform(df.CONTENT.astype(str))
df_eda['SENTIMENT'] = df.SENTIMENT
def show_dist(df, col):
print('Estadísticas descriptivas para el {}'.format(col))
print('-'*(len(col)+22))
print(df.groupby('SENTIMENT')[col].describe())
bins = np.arange(df[col].min(), df[col].max() + 1)
g = sns.FacetGrid(df, col='SENTIMENT', size=5, hue='SENTIMENT', palette="PuBuGn_d")
g = g.map(sns.distplot, col, kde=False, norm_hist=True, bins=bins)
plt.show()
show_dist(df_eda,'conteo_palabras')
show_dist(df_eda,'conteo_palabras_mayusculas')
# Se observa que no hay grandes diferencias entre la cantidad de palabras en las tres categorías de SENTIMENT, más del 75% de textos en todas las categorías tienen menos de 10 palabras.
# Observando la forma de la distribución se puede decir que los comentarios negativos, tienen más palabras al compararse con las otras dos categorías.
# Por lo menos la mitad de los comentarios neutrales tienen una palabra o ninguna.
# En el resto de clasificaciones (menciones, hashtags, palabras en mayúscula, signos de exclamación o interrogación, URLs y emojis) no se observan diferencias entre grupos y también se observa que muy pocos textos en CONTENT contienen alguna de las clasificaciones.
# Los comentarios no tienen menciones (palabras seguidas por '@'), hashtags ni URLs. Estos tienen pocos emojis, signos de interrogación ni exclamación y tampoco palabras en mayúscula. No se observan diferencias en estas clasificaciones entre las categorías de SENTIMENT.
# # Preprocesamiento
#
# A continuación se van a convertir todas las mayúsculas en minúsculas, se eliminaran las tildes, las URLs y los signos de puntuación.
## Vamos a eliminar las URL
df['CONTENT']=df['CONTENT'].astype(str)
df['CONTENT'] = df['CONTENT'].apply(
lambda x: re.sub(r'http\S+', '',x)
)
# +
# Convertimos todo el texto a minúscula
df['CONTENT'] = df.CONTENT.apply(lambda x: x.lower())
# +
## Reemplazar tildes y puntuación y cambiar algunas palabras
df['CONTENT'] = df['CONTENT'].apply(
lambda x: re.sub('á', 'a', x)
)
df['CONTENT'] = df['CONTENT'].apply(
lambda x: re.sub('é', 'e', x)
)
df['CONTENT'] = df['CONTENT'].apply(
lambda x: re.sub('í', 'i', x)
)
df['CONTENT'] = df['CONTENT'].apply(
lambda x: re.sub('ó', 'o', x)
)
df['CONTENT'] = df['CONTENT'].apply(
lambda x: re.sub('ú', 'u', x)
)
df['CONTENT'] = df['CONTENT'].apply(
lambda x: re.sub(r'[^\w\s]','',x)
)
df['CONTENT'] = df['CONTENT'].apply(
lambda x: re.sub('hassan','hassam',x)
)
df['CONTENT'] = df['CONTENT'].apply(
lambda x: re.sub('hasam','hassam',x)
)
# -
# # Frecuencia de las palabras en todo el dataset y en las diferentes categorías de SENTIMENT
texto=''
for i in df['CONTENT']:
texto += i+' '
texto=re.sub('hassan','hassam',texto)
tokens = nltk.tokenize.word_tokenize(texto)
pd.Series([w for w in tokens if len(w) > 3]).value_counts()
pd.Series([w for w in tokens if len(w) > 3]).value_counts()[:15][::-1].plot(kind='barh',title='Frecuencia de las palabras en todo el dataset')
texto=''
negativos=df[df['SENTIMENT']=='Negative']
for i in negativos['CONTENT']:
texto += i+' '
texto=re.sub('hassan','hassam',texto)
tokens = nltk.tokenize.word_tokenize(texto)
pd.Series([w for w in tokens if len(w) > 3]).value_counts()
pd.Series([w for w in tokens if len(w) > 3]).value_counts()[:15][::-1].plot(kind='barh',title='Frecuencia de las palabras en los comentarios negativos')
texto=''
neutrales=df[df['SENTIMENT']=='Neutral']
for i in neutrales['CONTENT']:
texto += i+' '
texto=re.sub('hassan','hassam',texto)
tokens = nltk.tokenize.word_tokenize(texto)
pd.Series([w for w in tokens if len(w) > 3]).value_counts()
pd.Series([w for w in tokens if len(w) > 3]).value_counts()[:15][::-1].plot(kind='barh',title='Frecuencia de las palabras en los comentarios neutrales')
texto=''
positivos=df[df['SENTIMENT']=='Positive']
for i in positivos['CONTENT']:
texto += i+' '
texto=re.sub('hassan','hassam',texto)
tokens = nltk.tokenize.word_tokenize(texto)
pd.Series([w for w in tokens if len(w) > 3]).value_counts()
pd.Series([w for w in tokens if len(w) > 3]).value_counts()[:15][::-1].plot(kind='barh',title='Frecuencia de las palabras en los comentarios positivos')
# En todas las categorías la palabra hassam es la más frecuente, esto muestra la popularidad de este comediante. Sin embargo, la mención de Hassam no parece estar asociada con ningún sentimiento en particular. Se observa lo mismo para el comediante Lokillo.
#
# En los comentarios negativos se encuentran palabras relacionadas con el desinterés o el aburrimiento con respecto a los comediantes y al programa.
#
# En los comentarios neutrales destaca la palabra empate y palabras relacionadas con votar.
#
# Las palabras más optimistas se encuentran en la categoría de sentimientos positivos, como por ejemplo, las palabras excelente o bueno. Las personas que escribieron estos comentarios estarían más involucradas con el programa y su contenido.
#
# # Clasificador Support Vector Machine con kernels lineal y rbf
# +
# Para poder usar los algoritmos de análisis de sentimientos, se deben eliminar las palabras conocidas como stopwords
# que son artículos o preposiciones que no aportan al sentido de los comentarios
stop = stopwords.words('spanish')
df["CONTENT"]=df["CONTENT"].apply(lambda words: ' '.join(word.lower() for word in words.split() if word not in stop))
df['CONTENT']
# +
# Se usan el 30% de los datos para validación y el 70% restante para el entrenamiendo
# Se codifican las clases de SENTIMENT para que sean enteros
asignacion_1 = {'Positive': 1, 'Negative': -1, 'Neutral': 0}
def decode_sentiment(label):
return asignacion_1[label]
df.target = df.SENTIMENT.apply(lambda x: decode_sentiment(x))
x_train, x_test, y_train, y_test = train_test_split(
df.CONTENT, df.target, test_size=0.3, shuffle=False, random_state=11)
# +
# Crear vectores de características, esto asigna un número a cada palabra de nuestro dataset
vectorizer = TfidfVectorizer(min_df = 5,
max_df = 0.8,
sublinear_tf = True,
use_idf = True)
train_vectors = vectorizer.fit_transform(x_train)
test_vectors = vectorizer.transform(x_test)
# +
# Clasificador SVM con kernel lineal
classifier_linear = svm.SVC(kernel='linear')
classifier_linear.fit(train_vectors, y_train)
prediction_linear = classifier_linear.predict(test_vectors)
report = classification_report(y_test, prediction_linear, output_dict=True)
print('Negativos:',report['-1'])
print('Neutrales:',report['0'])
print('Positivos:',report['1'])
# +
# Matriz de confusión
# La matriz de confusión nos indica qué tan bien clasificó nuestro algoritmo a los datos de validación
confusion = confusion_matrix(y_test, prediction_linear)
print('La matriz de confusión para el SVM con kernel lineal es: \n',confusion)
# +
# Clasificador SVM con kernel rbf
classifier_rbf = svm.SVC(kernel='rbf',gamma=0.3)
classifier_rbf.fit(train_vectors, y_train)
prediction_rbf = classifier_rbf.predict(test_vectors)
report = classification_report(y_test, prediction_rbf, output_dict=True)
print('Negativos:',report['-1'])
print('Neutrales:',report['0'])
print('Positivos:',report['1'])
# +
# Matriz de confusión
confusion = confusion_matrix(y_test, prediction_rbf)
print('La matriz de confusión para el SVM con kernel rbf es: \n',confusion)
# -
# ### Validación cruzada para el kernel lineal
#
# La validación cruzada nos permite probar nuestro clasificador en diferentes muestras aleatorias de los datos. Se crean $n$ muestras de entrenamiento y $n$ muestras de validación (en este caso escogemos $n=5$) de forma que todos los datos se encuentren una vez en la muestra de entrenamiento y una vez en la muestra de validación.
# Esto permite observar si el clasificador funciona bien en todos los datos o si el accuracy obtenido solo se tiene para los datos de entrenamiento y validación escogidos inicialmente.
clf_lineal = svm.SVC(kernel='linear')
train_vectors = vectorizer.fit_transform(df['CONTENT'])
cv_scores=cross_val_score(clf_lineal, train_vectors,df.target,cv=5)
print('Cross-validation (accuracy)', cross_val_score(clf_lineal, train_vectors,
df.target,
cv=5))
print('Promedio cross-validation score (5-fold): {:.3f}'
.format(np.mean(cv_scores)))
# ### Validación cruzada para el kernel rbf
clf_rbf = svm.SVC(kernel='rbf',gamma=0.3)
train_vectors = vectorizer.fit_transform(df['CONTENT'])
cv_scores=cross_val_score(clf_rbf, train_vectors,df.target,cv=5)
print('Cross-validation (accuracy)', cross_val_score(clf_rbf, train_vectors,
df.target,
cv=5))
print('Promedio cross-validation score (5-fold): {:.3f}'
.format(np.mean(cv_scores)))
# # Clasificador árboles de decisión
train_vectors = vectorizer.fit_transform(x_train)
classifier_random = DecisionTreeClassifier().fit(train_vectors, y_train)
prediction_random=classifier_random.predict(test_vectors)
report = classification_report(y_test, prediction_random, output_dict=True)
print('Negativos:',report['-1'])
print('Neutrales:',report['0'])
print('Positivos:',report['1'])
# +
# matriz de confusión
confusion = confusion_matrix(y_test, prediction_random)
print('La matriz de confusión para el SVM con kernel rbf es: \n',confusion)
# -
# ### Validación cruzada para el árbol de decisión
clf_random_cv = DecisionTreeClassifier()
train_vectors = vectorizer.fit_transform(df['CONTENT'])
cv_scores=cross_val_score(clf_random_cv, train_vectors,df.target,cv=5)
print('Cross-validation (accuracy)', cross_val_score(clf_random_cv, train_vectors,
df.target,
cv=5))
print('Promedio cross-validation score (5-fold): {:.3f}'
.format(np.mean(cv_scores)))
# Debido al desbalance que se presenta en los datos (hay muy pocos comentarios negativos comparados con los neutrales y los positivos), el f1 score es una buena métrica para evaluar el desempeño de los tres clasificadores.
# Los puntajes f1 para cada uno de los clasificadores son:
print('Accuracy del clasificador SVM con kernel lineal: {:.2f}'.format(accuracy_score(y_test, prediction_linear)))
print('Accuracy del clasificador SVM con kernel rbf: {:.2f}'.format(accuracy_score(y_test, prediction_rbf)))
print('Accuracy del clasificador árbol de decisión: {:.2f}'
.format(classifier_random.score(test_vectors, y_test)))
print('El f1 para el SVM con kernel lineal es: ',f1_score(y_test, prediction_linear, average='macro'))
print('El f1 para el SVM con kernel rbf es: ',f1_score(y_test, prediction_rbf, average='macro'))
print('El f1 para el árbol de decision es: ',f1_score(y_test, prediction_random, average='macro'))
# # Ejemplo de una predicción
#
# Observemos qué sentimiento asignan los tres clasificadores a un pequeño comentario
vectorizer = TfidfVectorizer(min_df = 5,
max_df = 0.8,
sublinear_tf = True,
use_idf = True)
train_vectors = vectorizer.fit_transform(x_train)
test_vectors = vectorizer.transform(x_test)
review_vector = vectorizer.transform(['un show aburrido en mi concepto no debería haber ningún ganador']) # vectorizing
print('Clasificación del SVM con kernel rbf:',classifier_rbf.predict(review_vector))
review_vector = vectorizer.transform(['un show aburrido en mi concepto no debería haber ningún ganador']) # vectorizing
print('Clasificación del SVM con kernel lineal:',classifier_random.predict(review_vector))
review_vector = vectorizer.transform(['un show aburrido en mi concepto no debería haber ningún ganador']) # vectorizing
print('Clasificación del árbol de decisión:',classifier_linear.predict(review_vector))
# Como vemos, se clasificó nuestro comentario como negativo (-1) en todos los algoritmos.
# # Conclusion
#
# Los tres clasificadores se ejecutan de forma rápida y tienen resultados aceptables, observando el accuracy promedio obtenido usando validación cruzada, el mejor algoritmo de los escogidos (medido por el accuracy) es el SVM con un kernel lineal.
# El SVM con kernel lineal tiene un mejor desempeño para clasificar comentarios neutrales, mientras que el kernel rbf tiene mejores resultados con los comentarios positivos. Dependiendo del análisis costo-beneficio que se haga (por ejemplo, si clasificar correctamente los comentarios neutrales resulta más beneficioso que clasificar de manera correcta las otras dos categorías) podría resultar mejor usar el SVM con kernel lineal a pesar de tener métricas ligeramente inferiores al kernel rbf.
# El SVM con kernel rbf se ajusta mejor que los otros dos algoritmos a la naturaleza desbalanceada de los datos, esto se concluye del f1 score, por lo que finalmente este podría ser el algoritmo con el mejor desempeño.
| 17,493 |
/Section 7 - Milestone Project - 1/Accepting User Input.ipynb | 748f8b413559f7a92cc78812296d998dd25420e7 | [] | no_license | Sejopc/Python3.0-Excercises | https://github.com/Sejopc/Python3.0-Excercises | 0 | 0 | null | null | null | null | Jupyter Notebook | false | false | .py | 4,845 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
result = input("Please enter a value: ")
result
type(result) # input function will always return a string
result = input("Please enter a value: ")
result = int(result)
type(result)
type(2.3)
float('3.14')
position_index = int(input("Choose an index position: "))
position_index
row1 = [1,2,3]
row1[position_index]
result = input("Enter a number: ")
2+2 # this will not work until we enter a value on above input. It is still waiting for user interacion for the input
# Also, if we run the input cell twice, the first iteration will still wait for the user input, which we are not able to provide because when we run the cell twice, it will erase the input box.
#
# Solution: Kernel tab > Restart
#
# However you will need to run ALL the cells again, as all variables, functions and stuff gets restarted.
#
# Quick way to do that:
#
# Cell > Run All (or Run All Above or Run All Below)
| 1,167 |
/NUMPY.ipynb | 78e459b4c7553e178282c9269d05c0fd4b98a219 | [] | no_license | MudassirRaza12345/ASSIMENTS | https://github.com/MudassirRaza12345/ASSIMENTS | 0 | 0 | null | null | null | null | Jupyter Notebook | false | false | .py | 44,965 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # READ FILE EXAMPLE START
# #Today Class Target
# How can we file and file manipulation,Libraries ko kaisa import krta ha
# Also we covered Numpy
# Numpy ka matlab means mukti dimensions array so array ka darmian ap samishan
# krata ha is normally done throug python programming ap loop lagana prta ha
# jis ka behalf pr wo proper kam krta but in numpy it can work in proper way
# numerical computing hogae,apka ps koi algebric ka behalf ma ap jb matrices
# ki koi implementation krta ho for that purpose we used numpy library
#
# +
#How can we read other file
#on home page New
# Notebook:
# python3
# Otherfile:
# Textfie
# Folder
# Terminal
#we go on Textfile create text file and save it without save it will not work.
#ab mna jo textfile banae ha is ka data ko ma pthon is page pr read kro
#ab mna with open("untitled.txt")apni file ka naam ps krdiya
#as ksi variable ka ander humko ya data chaya
#ab ma is data aik variable ma stored kro ga jis ka naam mna contents rkha ha
#ab mujha is pr read ka fuction perform krwana ha
#open ko jo method hota ha jo apki is file ko open kraga jo ap read krna chata
#ho
#then jo apka ps content araha isko specific name dena zarori ha then
#with ka keyword for example open ka keyword sa ap na file open to krdi
#but close kaisa kra ga to with ka function ya functionality perform krta
#jb ap ki file ma sa tamam content read hojata ha to with ka keyword
#automatically close krdega file ko
with open("untitled.txt") as practicefile:
contents= practicefile.read()
print(contents)
# -
#mna text file ma extra white space de ha
#123
#456
#789
#
#
#
#
#ab ma cha ta wo extra whitte space na print ho to hum apna code rstrip()
# used kra ga
#iski waja extra nahi ae gi
with open("untitled.txt") as practicefile:
contents= practicefile.read()
print(contents.rstrip())
# +
#ab chata hon aik folder bnao is folder sa mera data get ho
#How can we read other folder file
#on home page New
# Notebook:
# python3
# Otherfile:
# Textfie
# Folder
# Terminal
#we go on folder create folder
#ab ap nicha dekha ga untitled folder ab is pr click kra ga to ap ko ya
#options mila ga
# New
# Notebook:
# python3
# Otherfile:
# Textfie
# Folder
# Terminal
#ap text file pr click krka aik textfile bnalaga
#and save it without save it will not work.
#mera ps untitled ka naam sa aik file moujood ap ma isko naam
#doga untitled2
#mujha yha apna folder ka naam dena hoga iska baad folder jo file ha is ka naam
#dena hoga bqi syntax waisa hi rha ga jo textfile ka lya used kiya ha
with open("Untitled Folder/untitled2.txt") as practicefile:
contents= practicefile.read()
print(contents.rstrip())
# -
#abhi tq hum with open() ka ander file ps kr rha tha ab hum aik variable bna kr
#is ka file ps kra ga
#mna file path ka variable bnalya ha ab hum is ka ander aik dafa file location
#dalda ga is ka baad dobra dalna ki hum ko zarorat nhi
filepath = "Untitled Folder/untitled2.txt"
#is ka baad with open ap just file ps kra ga is ma location ko br br dena ki
# zarorat nhi hogi
with open(filepath) as practicefile:
contents= practicefile.read()
print(contents.rstrip())
# +
#agr ma chata hon for ka loop chalaao with ka method to ma asa kya karo ka
#tamam cheeza mujha for ka loop ma mil jaa with ka ander ki
#filepath = "untitled.txt"
#with open(filepath) as practicefile:
# contents= practicefile.read()
#ab ma chata ho yha print na ho balqa outside of the scoope print ho
#mera with scope and(khatam) ho chuka ha ab ma chata hon
#ab is scope sa bhr cheezon ko get kro to wo ma kaisa kro ga
#hum na loop ka ander forin ka loop pra tha ya kb kam ata ha jb text file
#ka ander multiple line ho ,jb multiple aka ps content ho to ap isko loop
#chalaka one by one kaisa get kra
#ab hum srf read konhi balqa readlines ka method ko used kra ga
filepath = "untitled.txt"
with open(filepath) as practicefile:
contents= practicefile.readlines()
for line in contents:
print(line)
# +
#for removing empty spaces we rstrip()
filepath = "untitled.txt"
with open(filepath) as practicefile:
contents= practicefile.readlines()
for line in contents:
print(line.rstrip())
# +
#ab ma chta ho mera jitna data ha wo alag line ma print hona ka bjaa aik single
#line ma print hojaa to
# +
filepath = "untitled.txt"
with open(filepath) as practicefile:
contents= practicefile.readlines()
#first we declare empty variable
oneline=""
for line in contents:
oneline += line.rstrip() #ab hum .rstrip used kra ga jis answer aik
# line ma print hojaa ga or agr hum rstrip ka method ko call nhi krta to
#wo empty space ko neglect nhi kra ga
#oneline += line means oneline aik line pr print kra .
# #+= apna jo cheez pass ki ha wo iska ander hi append(add)krta rha ga
#one file ma line sa value mila gi wo brbr add hota rha gi isma or jakr
#phir oneline"" stored hojaa gi.
print(oneline)
# +
#ab ap chta ha ka apki file sab content na ae balqa specific content ae
#for 123456789 ma 123456 show ho is baad koi content show na ho
filepath = "untitled.txt"
with open(filepath) as practicefile:
contents= practicefile.readlines()
#first we declare empty variable
oneline=""
for line in contents:
oneline += line.rstrip()
#slicing ka method used kra ga
#[:]
#[:6]agr hum isma starting ma kuch nhi likha ga to phela
#number pr cheez hogi isko lala ga first pr
#like yha one ha is lya one is jga pr rekla ga
print(oneline[:6] +" "+"still continue")
#ab hum check krta ha slicing krna ka baat iski initial length baqi rhagi
# ya nhi
print(len(oneline))
#to iski len isna 9 hi bataae change nhi hoe
print("\n------------------------ xxxxxx-----------------------\nExamples of file read end\n------------------------ xxxxxx-----------------------")
# +
#ab ma chata ho ka ma apni file ka ander koi cheez write krwada
#yan koi code likho ya koi line likho jo file ma jaka stored hojaa
filepath = "untitled.txt"
with open(filepath,'w') as practicefile:
practicefile.write("000")
#agr apko write ka operation perform krna ha to ap filepath ka sath "w" likha
#ga jo write ka operation ko bata rhi ha.
# +
filepath = "untitled.txt"
with open(filepath,'w') as practicefile:
practicefile.write("000\n")# \n when use multi-line
# PracticeFile.write("111") #for multi-line for separation we used \n
practicefile.write("111\n")
# +
#jb mna write ka operation perform kiya to mera ps prona data remove ho gya
#or new data jo mna write ka zarya diya wo agya ab ma chata hon kuch asa kro
#ka porana data bhi mujha miljaa.
#or ma jo new cheez likhwana chahrha wo print ho
#iska hum aik operation perform krta jisko append ka operation kheta ha
filepath = "untitled.txt"
#agr ma yha write ka operaion perform krta hon to content ma overwrite krdaga
#or porana data(content)ko removed krdaga islya hum na 'a'(append) likha jis
#wo is file ma ajaaga or porana data bhi rha ga
with open(filepath,'a') as practicefile:
practicefile.write("222")
# +
#suppose we 5 divide 0 agr ya kaam python ko bolo ga to wo dega 0 division
#error to hamara ps python ka ander jo cases hota hainko kheta ha exception
#agr in ko control krna ha jaise ap error pta ha koi bhi cheez 0 sa divide
#to error ae ga 0 division error
#ab hum maaloom python error dedaga to ya cheez humko execution sa phela check
#krani ha
#All programming language ka ander try and catch ko used krta ho
#try ma koi cheez ko try krta ha
#or catch ma isma agr koi error hota ha to check krta ha
#python ka ander hamara ps hota try or exception ka block hota
#koi cheez ap try krta ho suppose in do ko divide krta ho
#exception ma jo error ae iska against ma apna kya answer dena wo batana hoga
firstnum=5
secondnum=0
try:
answer=firstnum/secondnum
except ZeroDivisionError:
print("This operation cannot perform")
else:#agr zero na ho to is else ka block ma answer ko print krdo
print(answer)
# +
firstnum=5
secondnum=3
try:
answer=firstnum/secondnum
except ZeroDivisionError:
print("This operation cannot perform")
else:#agr zero na ho to is else ka block ma answer ko print krdo
print(answer)
# +
#some time you find some file python give error that file not found error
#agr is ko file na mila
try:
with open("untitled.txt") as practicefile:
contents= practicefile.read()
#agr isko file nhi mile
except FileNotFoundError:
print("your file path is not correct")
else:#agr file present ho to tamam cheeza contents ma ajaa gi
print(contents)
# +
#some time you find some file python give error that file not found error
#agr is ko file na mila
try:
with open("untitled3.txt") as practicefile:
contents= practicefile.read()
#agr isko file nhi mile
except FileNotFoundError:
print("your file path is not correct")
else:#agr file present ho to tamam cheeza contents ma ajaa gi
print(contents)
# -
# # MODULE EXAMPLE START
#ap ka excel ki file hoti ha wo CSV(Comma Separated Value) file hoti ha
#ab agr ap ka ps koi excel ki file ha isma muliple coloumns hota ha
#multiple data hota ha in file ka ander ap data kaise get kro ga
#suppose excel ki file is ma sa ap data kaise get kro ga
#sab sa apna csv ka module import krna
import csv
with open("Book2.csv") as practicefile:
contents= csv.reader(practicefile)
for line in contents:
print(line)
#csv.reader is the syntax to excess
#csv file ab for ka loop ki zarorat paragi to excess row and coloumn in excel
#file
# +
import csv#EXPORT AS CSV
with open('data.csv') as PracticeFile:
contents=csv.reader(PracticeFile)
for line in contents:
print(line)
# -
#BULTIN MODULE HA HAMRA PS HA MATH KA MODULE TO HUM MODULE KO IMPORT KRTA HA
import math#hum ab math ka tamam module ko import krsakta ha
print(math.pi)
#ab ma apsa kheta ha ka math ka jo module arha is ka naam change krdo
import math as mymath#ab ma module apni tamam file mymath ka zarya imort krloga
#ab mujha agr pi ki value chaya ha to mymath ka zarya print krlo ga
print(mymath.pi)
#ap ka math ka jo module ha is ma bhot sara operation ha like pi ka ha, square
#ka ha,ceiling,floor,log etc iska multiple operation ha apko pora module import
#nhi krna is ma sa ksi specific fuction ko import krna ha
from math import pi
print(pi)
from math import * #star ka matlab ha is module ki tamam cheeza import krlo ga
print(pi)
import math #ceiling means next value (or ya value ko round off krka increase
#krdeta ha)
math.ceil(5.7)
import math #floor means previous value(ya round off kr value ko degrade krdeta
#ha yani kam kr deta ha )
math.floor(5.7)
# +
#ab mna aik new python ki file banae ha whan mna aik fuction bnaya ha
#def add(a,b):
# result = a+b
# return result
#ap is file ka ander function ka result yha hamrae module file ma excess
# kr sakta ha
#ab is tarh ka module hum is tarh import krta ha
import import_ipynb#ab ma jb import_ipynb likho ga to ma insure krdo ga ka wo
#wo inma sa ksi file ko import krla
#ab yha sab sa phela apni file ko import kro ga
import Untitled1
#Untitled1. ka sath jo function perform krna is ko likho ga
Untitled1.add(5,3)
# +
#ab mna aik new python ki file banae ha whan mna aik fuction bnaya ha
#def add(a,b):
# result = a+b
# return result
#ap is file ka ander function ka result yha hamrae module file ma excess
# kr sakta ha
#ab is tarh ka module hum is tarh import krta ha
import import_ipynb#ab ma jb import_ipynb likho ga to ma insure krdo ga ka wo
#wo inma sa ksi file ko import krla
#ab yha sab sa phela apni file ko import kro ga
import Untitled1
#Untitled1. ka sath jo function perform krna is ko likho ga
Untitled1.add(5,3)
#error is lya aya tha hum ko ipynb ka jo module ha is install krna zaroori ha
#ab ma jb isko install kro ga to error gaib hojaa ga
#ab ab home pr gae home pa ana ka baad new
# Notebook
# python3
# other
# TextFile
# Folder
# Terminal
#ab hum terinal p jae ga wha ya (pip install import_ipynb)(ya command is baat
#ko insure kr rhi ha ka aik file ko dosri file ma excess krsaktaha)
#ya command likho ga #or module install ho jaa ga phir upper wala
#programrun ho jaa ga
# -
# # NUMPY STARTING
# #INTRODUCTION:
# Numpy apko functionality deta ha multi dimension array bnaa ki
# is ka do package or ha 1)Numpy 2)pandas
# ya basically fundamental or bhot important package ha apki pyhton programming
# ka kyu ya is cheez ko insure krta apka deta multidimension store krta ha
# ap ka apka complex operation jo perform horha hota jis ma apka linear algebra,
# computational counting ya tamam ki tamam apka ps jo cheeza perform hoti ha
# bhot huge calculation hoti ha to wha pr ap python ki list or touple wagira
# sa kra ga to bhot musqil sa hoga ap isko control nhi krsakta ha
# inko apko controll krna la lya Numpy,pandas in frame work utilize krta ha
# apko isko utilize to kr leta ha is baat kya hota ha
# ap agr do array ko add krna chta ha to isma ap index add no ko add kra ga
# arr1[1,2,3] and arr2[4,5,6] agr ap indono ko add kro ga to for ka loop apply
# kro ga python jb ka numpy ma ap by the help of plus(+) sign ap do arrays
# ko add krdeto ho
# ap ka brae sa brae array ho muti dimension ho ap iska + sign ki madad sa add
# krsakta ho or linear algebra or complex calculation ma apko functionality
# provide krta ha ka ap apna data asani sa manipulate krsakta ha or used krsakta
# ha
# multidimensionarray means aik ziada array
#
#ab br numpy likna ka bjae mna isko np sa initialize kr dya ab brbr numpy nhi
#likha ga just np likha ga
import numpy as np
#now we generate random number we write np.random.randn likha ga numpy ma to wo
#mujha random number generate krda ga
#ab mna jo (2,3) likha ha is ma 2 ka matlab ya ha apko do array chaya or ,3
#ka matlab ya ha ka apko hr array ka ander 3 random number chya ha
data = np.random.randn(2,3)
print(data)
# +
import numpy as np
data = np.random.randn(2,3)
print(data,'\n')
print(data+data)
#ab ma charha hon data ka ander jo cheez generate hue ha wo dobra isi kaander
#add hojaga
# +
import numpy as np
data = np.random.randn(2,3)
print(data,'\n')
print(data+data,'\n')
print(data * 10)#data ka ander jitna bhi number ha inko multiply krdo 10 sa
# +
import numpy as np
data = np.random.randn(2,3)
print(data,'\n')
print(data+data,'\n')
print(data * 10,'\n')
print(data.shape)#data.shape mujha ya btata ha ap ka ps kitni arrays ha or is
#ma kitna number ha
# +
import numpy as np
data = np.random.randn(2,3)
print(data,'\n')
print(data+data,'\n')
print(data * 10,'\n')
print(data.shape,'\n')
print(data.dtype,'\n')#dtype hamra ps data type btata ha ka hamara data kis type
#ka ha
# +
import numpy as np
data = np.random.randn(2,3)
print(data,'\n')
print(data+data,'\n')
print(data * 10,'\n')
print(data.shape,'\n')
print(data.dtype,'\n')
print(data.ndim,'\n')#n means number or dim means dimension ka hara ps
# kitni array generate hue ha
# +
#Numpy apko functionality deta ha ap zero ki array bhi generate kr sakta ha
print(np.zeros(10))#humna aik jga numpy ko initialize krdiya ab hum hr jga
#np ka zarya np ko used krsakta ha
#10 represent no of zero
# -
print(np.zeros(10),'\n')
print(np.zeros((3,6)),'\n')#ab humna 3 arry bnae or is ma 6 zeros dala
print(np.zeros(10),'\n')
print(np.zeros((3,6)),'\n')
print(np.zeros((2,3,6)),'\n')#ab yha isna 2 array generate kiya iska ander
#3 array or banay or is ma 6 zero dal diya
print(np.zeros(10),'\n')
print(np.zeros((3,6)),'\n')
print(np.zeros((2,3,6)),'\n')
#isi trh hamra numpy ma empty ka module ab is ko dektha wo kaisa used hota ha
print('it is empty example \n',np.empty((3,6)),'\n')
#ab yha empty or zero ma ya difference ha ka empty apko hr 0 daga or kabhi apko
#garbage value daga jiska matlab koi bhi value
#to zaroori nhi ha is lya agr apko zero ki need ha to ap np.zeros print kra
#ab yha arra bnae ha ab is normal arry ko numpy ki arry bnae ga hum
arr = np.array([1,2,3,4,5,6])#ab yha ma jo bhi value doga wo numpy
#ki array bn jaage
#ab ma chata hon ya value jo int ma ha wo float ma convert hojaa iskaly
#hum used kra ga astype() or bracket ka ander jis ma convert krna ha wo
#likh da suppose float ka np.float
print(arr.astype(np.float64))
#means isna aik integer array ko float ma convert krdiya ha
arr = np.array([1,2,3,4,5,6])
print(arr.astype(np.float64))
arr = np.array([4,2,3,6,9],dtype='int64')#hum array bnata waqt iski data type bhi initialize
#kr sakta ha
print(arr *2)
arr = np.array([4,2,3,6,9],dtype='int64')
print(arr,'\n')
print(arr *2,'\n')
arr = np.array([4,2,3,6,9],dtype='int64')
print(arr,'\n')
print(arr *2,'\n')
print(arr * arr,'\n')
arr = np.array([4,2,3,6,9],dtype='int64')
print(arr,'\n')
print(arr *2,'\n')
print(arr * arr,'\n')
print(2 * arr,'\n')
print(arr ** 2,'\n')#**(for power)
print(np.sqrt(arr),'\n')# for square root
print(np.exp(arr),'\n')#for exponent
print(np.mean(arr),'\n')#Statistics operation (mean,median and mode)
print(np.sum(arr),'\n')#sum
print(np.cumsum(arr),'\n')#cumsum yani 4 ko 2 ma add kiya phir iska baad indono
#ka result ko next value ma add kiya like 3 ma kiya and so on
print(np.sort(arr),'\n')#sort(arrange)
# now we do boolean operation
#> <
#mujha check krna arry1 ki value arr2 ki value sa bari ha ya nhi
arr2 =np.array([0,1,3,5,4])
print(arr > arr2)
print(arr < arr2)
# +
#ab humko range ka function used krta python ma but numpy ma hum arange used
#krta ha
arr = np.arange(10)#ab mna arange 0 to taq ki value ko arrange bhi kya
#or generate bhi
print(arr)
arr2=arr[5:8]
print(arr2)
arr[:3]=0 #jin element ko slicing ki in tamam ko zero sa replace krdega ga
print(arr)
# -
namesArr = np.array(["ali","raza","fahad","hussain","ali"])
#jitna element first list ma hoga itna array generate kra
# second arr ma means 5 (5,3)
numArr = np.random.randn(5,3)
print(numArr ,'\n')
print(namesArr ,'\n')
print(namesArr == "ali",'\n')
print(numArr[namesArr == "ali"],'\n')#numArr ka ander check kro kya cheez ka ali apko
#apko kha kha mil rha ha jha jha ap ko ali mila is arr ma sa wo wala arr ko
#(value) ko uthao or print krdo to isna first wali arry or last lakr print kr di
#jha isko ali mila
print(numArr[~(namesArr == "ali")],'\n')#~ not ka operation
#ab 3 jga ali nhi tha islya inteen jga print kiya
print(np.unique(namesArr),'\n')#hr element ko print krddga jo repeat
#horha ho aik dafa print krda ga or sath ma sorting bhi krda ga(A,F,H,R)
import numpy as np
arr1 =np.random.randn(4)
print(arr1,'\n')
arr2 =np.random.randn(4)
print(arr2,'\n')
print(np.maximum(arr1,arr2),'\n')
print(np.minimum(arr1,arr2),'\n')
| 20,352 |
/5조_프로젝트_데이터확장_0614.ipynb | 12278660ecfbff918b1a4e0fa916df464d86a896 | [] | no_license | Deepfull/BootCamp | https://github.com/Deepfull/BootCamp | 0 | 0 | null | null | null | null | Jupyter Notebook | false | false | .py | 225,075 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# + [markdown] id="view-in-github" colab_type="text"
# <a href="https://colab.research.google.com/github/Deepfull/BootCamp/blob/main/5%EC%A1%B0_%ED%94%84%EB%A1%9C%EC%A0%9D%ED%8A%B8_%EB%8D%B0%EC%9D%B4%ED%84%B0%ED%99%95%EC%9E%A5_0614.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# + [markdown] id="sM9EAxW-oCCB"
# - 데이터확장
# + id="8B2q9YpA9etK"
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
# %matplotlib inline
# %config InlineBackend.figure_format = 'retina'
# + id="22WtGOUwMXZ2"
# 데이터 전처리
# add features 예제
# moving average
# window 정함 50
# 3sigma 정도로 limit 검
def data_processing(case):
num = case['num']
filter = case['filter']
circulation = case['circulation']
purifier = case['purifier']
# Github에서 데이터 읽기
path = 'https://raw.githubusercontent.com/eupshin/BootCamp/main/'
df_ca = pd.read_csv(path + 'ca' + num + '.csv')
df_oa = pd.read_csv(path + 'oa' + num + '.csv')
# CA,RA 데이터 정리
size_ca = df_ca.shape[0]
#print(num, ': ', size_ca)
df_ca['time']=list(range(0,size_ca))
df_ca = df_ca.drop('time1', axis=1)
# OA 데이터 정리
size_oa = df_oa.shape[0]
#print(num, ' :', size_oa)
#df_oa = df_oa[['OAPM10','OAPM2.5', 'OAPM1']]
df_oa = df_oa.drop('time2', axis=1)
df_oa10 = pd.DataFrame(columns=['OAPM10','OAPM2.5', 'OAPM1'])
for j in range(0,size_oa):
for i in range(0,10):
df_oa10.loc[i+j*10] = df_oa.loc[j][['OAPM10','OAPM2.5', 'OAPM1']]
# 데이터(CA,RA,OA) 합치기
df_con = pd.concat([df_ca,df_oa10],axis=1)
# 운전조건(필터) 추가
if filter == 'esp':
df_con['filter_esp'] = 1
df_con['filter_merv'] = 0
else:
df_con['filter_esp'] = 0
df_con['filter_merv'] = 1
# 운전조건(순환) 추가
if circulation == 'ex':
df_con['circulation_ex'] = 1
df_con['circulation_in'] = 0
else:
df_con['circulation_ex'] = 0
df_con['circulation_in'] = 1
# 운전조건(공청기) 추가
if purifier == 'off':
df_con['purifier_off'] = 1
df_con['purifier_on'] = 0
else:
df_con['purifier_off'] = 0
df_con['purifier_on'] = 1
return df_con
case11 = {'num':'1_1', 'filter':'esp', 'circulation':'ex', 'purifier':'off'}
df11 = data_processing(case11)
case12 = {'num':'1_2', 'filter':'esp', 'circulation':'ex', 'purifier':'on'}
df12 = data_processing(case12)
case21 = {'num':'2_1', 'filter':'esp', 'circulation':'in', 'purifier':'off'}
df21 = data_processing(case21)
case22 = {'num':'2_2', 'filter':'esp', 'circulation':'in', 'purifier':'on'}
df22 = data_processing(case22)
case32 = {'num':'3_2', 'filter':'merv', 'circulation':'ex', 'purifier':'off'}
df32 = data_processing(case32)
case41 = {'num':'4_1', 'filter':'merv', 'circulation':'in', 'purifier':'off'}
df41 = data_processing(case41)
case42 = {'num':'4_2', 'filter':'merv', 'circulation':'in', 'purifier':'on'}
df42 = data_processing(case42)
# + colab={"base_uri": "https://localhost:8080/", "height": 312} id="GLRam6WYr-9q" outputId="c7680577-f070-444b-afec-963c165f0b52"
plt.figure(figsize=(14,4))
df_p1 = df11
plt.subplot(1,2,1)
plt.plot(df_p1['time'], df_p1['CAPM10'])
plt.plot(df_p1['time'], df_p1['CAPM2.5'])
plt.plot(df_p1['time'], df_p1['CAPM1'])
plt.title('HVAC_On')
plt.legend(['PM10','PM2.5','PM1'])
plt.xlabel('Time step')
plt.ylabel('Concentration')
df_p2 = df12
plt.subplot(1,2,2)
plt.plot(df_p2['time'], df_p2['CAPM10'])
plt.plot(df_p2['time'], df_p2['CAPM2.5'])
plt.plot(df_p2['time'], df_p2['CAPM1'])
plt.title('HVAC_Off')
plt.legend(['PM10','PM2.5','PM1'])
plt.xlabel('Time step')
plt.ylabel('Concentration')
# + colab={"base_uri": "https://localhost:8080/", "height": 286} id="HVhxcI39jyK2" outputId="ad87d787-2999-4e8c-9268-5eff4924449c"
# 이동평균을 이용하여 피크값 제거
def rolling_average(df, window):
df_av = df.rolling(window, min_periods=1).mean()
df_av.columns = [col + "_av" + str(window) for col in df.columns]
return df_av
x = rolling_average(df11[['CAPM10']], 6);x
plt.plot(df11['CAPM10'].values)
plt.plot(x)
plt.legend(['df','x'])
# + colab={"base_uri": "https://localhost:8080/"} id="_btqOJgMkPNt" outputId="4203347b-aedc-460f-c66b-23e1dfbd62a3"
df11['CAPM10'].shape
# + id="Y-5CZ3D_acuS"
# t = t0일 때의 농도 값으로 t = t0 + 100일때의 농도 값을 예측
# 학습데이터
# X y
# data @ t=0 data @ t=10
# data @ t=1 data @ t=11
# data @ t=2 data @ t=12
# data @ t=3 data @ t=13
# 타임 스텝
time_step = 100
# 현재에서 몇% 떨어졌는지 추정
# shift할 때
def create_xy(df):
size_df = df.shape[0]
df_x = df[['CAPM10', 'CAPM2.5', 'CAPM1',
'OAPM10', 'OAPM2.5', 'OAPM1',
'filter_esp', 'filter_merv',
'circulation_ex', 'circulation_in',
'purifier_off', 'purifier_on']][:size_df-time_step]
#df_y = df[['CAPM10', 'CAPM2.5', 'CAPM1',
# 'RAPM10', 'RAPM2.5', 'RAPM1']][time_step:size_df]
df_y = df[['CAPM10','CAPM2.5', 'CAPM1']][time_step:size_df]
return df_x, df_y
# 학습 데이터 합치기
df_x, df_y = create_xy(df11)
df_x1, df_y1 = create_xy(df12)
df_x = pd.concat([df_x, df_x1])
df_y = pd.concat([df_y, df_y1])
df_x1, df_y1 = create_xy(df21)
df_x = pd.concat([df_x, df_x1])
df_y = pd.concat([df_y, df_y1])
df_x1, df_y1 = create_xy(df22)
df_x = pd.concat([df_x, df_x1])
df_y = pd.concat([df_y, df_y1])
df_x1, df_y1 = create_xy(df32)
df_x = pd.concat([df_x, df_x1])
df_y = pd.concat([df_y, df_y1])
df_x1, df_y1 = create_xy(df41)
df_x = pd.concat([df_x, df_x1])
df_y = pd.concat([df_y, df_y1])
df_x1, df_y1 = create_xy(df42)
df_x = pd.concat([df_x, df_x1])
df_y = pd.concat([df_y, df_y1])
#df_x.shape, df_y.shape
# + id="EC4LkCjOakSZ"
from sklearn.model_selection import train_test_split, cross_val_score
from sklearn.ensemble import RandomForestRegressor
# train, test 데이터 분할
X_train, X_test, y_train, y_test = train_test_split(df_x, df_y, random_state=9)
# 데이터 형식 변환
X_train = np.array(X_train)
X_test = np.array(X_test)
y_train = np.array(y_train)
y_test = np.array(y_test)
# 학습 실시
rfr = RandomForestRegressor(n_estimators=20)
rfr.fit(X_train, y_train)
rfr.score(X_test, y_test)
# + id="qE1YBSfsyXqn"
# 실측, 예측 비교
y_pred = rfr.predict(X_test)
plt.figure(figsize=(7,6))
plt.scatter(y_test[:,0],y_pred[:,0])
plt.scatter(y_test[:,1],y_pred[:,1])
plt.scatter(y_test[:,2],y_pred[:,2])
plt.title('CA : PM10, PM2.5, PM1')
plt.legend(['PM10','PM2.5','PM1'])
plt.xlabel('y_test')
plt.ylabel('y_pred')
plt.plot([0,100],[0,100],'--')
# + id="rx8z_KJ95Cma"
# t0 에서의 값으로 (t0+1) ~ (t0+10) 사이의 값을 예측
# 학습데이터
# X y
# data @ t=0 data @ t=1~10
# data @ t=1 data @ t=2~11
# data @ t=2 data @ t=3~12
# data @ t=3 data @ t=4~13
#
# 만약에 t = (t0+1) ~ (t0+20) 사이의 값을 예측하고 싶다면
# 위의 학습으로 예측된 t0+10값을 X로 입력하여 다음 10개 time step 값을 예측함
# Sequential regression
# Linear Model(여러번 돌려야됨), MLP
# LSTM을 통한 시계열 예측, MLP의 depth가 있음
mates_dic.get(element.Cabin) ) if (str(element.Cabin)!='nan') else 0,axis=1)
data_set['Roomates'].describe()
grid_searchgb.fit(X_train,survived_train)
grid_searchgb.best_score_
grid_searchgb.best_params_
grid_searchxgb.fit(X_train,survived_train)
grid_searchxgb.best_score_
grid_searchxgb.best_params_
rf=RandomForestClassifier(n_estimators=150, max_depth=6).fit(X_train,survived_train)
gb=GradientBoostingClassifier(n_estimators=10).fit(X_train,survived_train)
xgb=XGBClassifier(learning_rate=0.05,max_depth=2,n_estimators=50).fit(X_train,survived_train)
from sklearn.ensemble import VotingClassifier
vote=VotingClassifier(estimators=[('rf',rf),('gb',gb),('xgb',xgb)],voting='soft',weights=[1,1,1])
vote_final=vote.fit(X_train,survived_train)
survived_predictionv=vote.predict(y_test)
submission=pd.DataFrame({"PassengerID":y_test['PassengerId'],"Survived":survived_prediction})
submission.to_csv('vote4.csv',index=False)
data_set['Ticket'].describe()
data_set['Ticket'].value_counts()
| 8,024 |
/Pruebas Cortas/Prueba Corta 4/PruebaCorta4.ipynb | 8cca182f9ff5866e51d4f2f7c059e589ce9417be | [] | no_license | plasmallan/an-lisis_procesos_ii | https://github.com/plasmallan/an-lisis_procesos_ii | 0 | 0 | null | null | null | null | Jupyter Notebook | false | false | .py | 3,970 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Python
# language: python
# name: conda-env-python-py
# ---
import numpy as np # useful for many scientific computing in Python
import pandas as pd # primary data structure library
# +
# !conda install -c conda-forge folium=0.5.0 --yes
import folium
print('Folium installed and imported!')
# -
world_map = folium.Map()
world_map
# +
world_map = folium.Map(location=[56.130, -106.35], zoom_start=4, tiles='Stamen Toner')
# display map
world_map
# +
# create a Stamen Toner map of the world centered around Canada
world_map = folium.Map(location=[56.130, -106.35], zoom_start=4, tiles='Stamen Terrain')
# display map
world_map
# +
# create a world map with a Mapbox Bright style.
world_map = folium.Map(tiles='Mapbox Bright')
# display the map
world_map
# +
df_incidents = pd.read_csv('https://s3-api.us-geo.objectstorage.softlayer.net/cf-courses-data/CognitiveClass/DV0101EN/labs/Data_Files/Police_Department_Incidents_-_Previous_Year__2016_.csv')
print('Dataset downloaded and read into a pandas dataframe!')
# -
df_incidents.head()
limit = 100
df_incidents = df_incidents.iloc[0:limit, :]
latitude = 37.77
longitude = -122.42
# +
sanfran_map = folium.Map(location=[latitude, longitude], zoom_start=12)
# display the map of San Francisco
sanfran_map
# +
# instantiate a feature group for the incidents in the dataframe
incidents = folium.map.FeatureGroup()
# loop through the 100 crimes and add each to the incidents feature group
for lat, lng, in zip(df_incidents.Y, df_incidents.X):
incidents.add_child(
folium.features.CircleMarker(
[lat, lng],
radius=5, # define how big you want the circle markers to be
color='yellow',
fill=True,
fill_color='blue',
fill_opacity=0.6
)
)
# add incidents to map
sanfran_map.add_child(incidents)
# +
# instantiate a feature group for the incidents in the dataframe
incidents = folium.map.FeatureGroup()
# loop through the 100 crimes and add each to the incidents feature group
for lat, lng, in zip(df_incidents.Y, df_incidents.X):
incidents.add_child(
folium.features.CircleMarker(
[lat, lng],
radius=5, # define how big you want the circle markers to be
color='yellow',
fill=True,
fill_color='blue',
fill_opacity=0.6
)
)
# add pop-up text to each marker on the map
latitudes = list(df_incidents.Y)
longitudes = list(df_incidents.X)
labels = list(df_incidents.Category)
for lat, lng, label in zip(latitudes, longitudes, labels):
folium.Marker([lat, lng], popup=label).add_to(sanfran_map)
# add incidents to map
sanfran_map.add_child(incidents)
# +
sanfran_map = folium.Map(location=[latitude, longitude], zoom_start=12)
# loop through the 100 crimes and add each to the map
for lat, lng, label in zip(df_incidents.Y, df_incidents.X, df_incidents.Category):
folium.features.CircleMarker(
[lat, lng],
radius=5, # define how big you want the circle markers to be
color='yellow',
fill=True,
popup=label,
fill_color='blue',
fill_opacity=0.6
).add_to(sanfran_map)
# show map
sanfran_map
# +
from folium import plugins
# let's start again with a clean copy of the map of San Francisco
sanfran_map = folium.Map(location = [latitude, longitude], zoom_start = 12)
# instantiate a mark cluster object for the incidents in the dataframe
incidents = plugins.MarkerCluster().add_to(sanfran_map)
# loop through the dataframe and add each data point to the mark cluster
for lat, lng, label, in zip(df_incidents.Y, df_incidents.X, df_incidents.Category):
folium.Marker(
location=[lat, lng],
icon=None,
popup=label,
).add_to(incidents)
# display map
sanfran_map
# -
| 4,035 |
/python/007_Pandas_200107.ipynb | 93609a8c3e14c3b91b92f25a83ceea15e02d9213 | [] | no_license | leeyunji964/Yunji | https://github.com/leeyunji964/Yunji | 0 | 0 | null | null | null | null | Jupyter Notebook | false | false | .py | 1,180,113 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# +
# 미분:
# 분산 : (sigma(관측치 - 평균)^2) / n-1
# 표준편차 : root((sigma(관측치 - 평균)^2) / n-1)
# 공분산 : sigma(x-xbar)(y-ybar) / n-1
# 상관계수 : (sigma (x-xbar)(y-ybar) / n-1) / (xsigma * ysigma) : -1 ~ 1
# 벡터 거리값 : 각 요소의 차의 제곱을 루트로 구함
# model 평가 : MSE(mean square error) : 연속된 수치인 경우의 평가
# RMSE(root)
# 신경망에서 cost function 비용함수 : 예측치 - 관측치 => back propagation( 역전파를 통해서 가중치를 수정)
# 국어, 수학 점수 : vector : 상관계수가 높다 -> 두 변수 간의 관계가 있다.
# (내적 = 1 ) = (상관계수 = 1) 둘이 같은 의미 : 상관계수가 구하기 더 복잡하니까 내적 사용.
# -
# ## 회귀분석, 상관분석
# - 회귀분석 다중공선성 : 종속변수가 과하게 영향을 받을 수 있다.
# - 회귀분석에서 자기상관성(Auto corelation)이 존재한다면? -> 시계열 분석을 해야한다.
# - 시계열 분석 ARMA ( Autocorelation + Moving Average)
# - 이동평균법 ( 주파수를 줄여준다 -> 복잡하면 분석 불가 )
# - 정상성 데이터에만 적용 가능 :평균이 일정하고 분산이 일정한.. 그런..데이터
# - 비정상 -> 정상성으로 변환
# - ARIMA ( integrated 통합 => 비정상성을 띈 데이터에도 가능)
# - pandas는 시계열 분석을 지원 ( 시간 index를 지원)
#
# +
import numpy as np
from statistics import *
x = np.array([7,8,9])
y = np.array([9,10,20])
print(y.mean()) # 통계에서는 mean을 안 쓰고 중위수(median)로 사용
# median을 사용하는 이유 : 이상치의 영향을 받지 않기 때문.
# 공분산 행렬
print("공분산", np.cov(x,y))
# 상관계수 행렬( 열 2, 행 2 : 4 )
print("상관계수", np.corrcoef(x,y))
# -
import numpy as np
arr = np.array([[1,2,3],[4,5,6],[7,8,9]])
print("원본", arr)
print("누적합(행)=", arr.cumsum(0))
print("누적곱(열)=", arr.cumprod(1))
print("배열의 합계 = ", arr.sum())
print("열방향으로의 합계(행평균) =", arr.mean(axis=1)) # 행방향과 열방향의 구분
print("행방향으로의 합계=", arr.sum(0)) # 열별 합계
# +
# 가중치( 변수 중요도를 부여)를 부여한다는 의미
a = np.array([1,2,3,4])
wts = np.array([4,3,2,1])
print( np.average(a, weights = wts)) # weights = 가중치 (수의 중요성 ) = wts
std = np.sqrt(np.mean(abs(a - a.mean())**2))
print(np.var([1,2,3,4])) #분산
print(np.std([1,2,3,4])) # 표준편차
# -
# # matplotlib, seaborn, pandas.plot
# - matplotlib로 구성
# - %matplotlib inline을 사용해야 출력됨
# +
# %matplotlib inline
from numpy import mean, std # 메모리낭비를 절감
#from numpy import std
from numpy.random import randn # 서브 패키지 : linalg(선형계수), random, fft(푸리에 변환), poly(다차방정식)
from numpy.random import seed
# seed 값을 주는 이유 : 컴퓨터에서 random은 의사랜덤(난수) - 랜덤처럼 보이지만 실은 데이터 출력 순서가 있는 것
# seed는 그 시작점을 항상 같게 해줌 -> 랜덤인데 같은 수가 나오게 한다.
from matplotlib import pyplot
seed(1) # 시작점은 1
data1 = 20 * randn(1000) + 100 # randn = randnormal : 정규분포에서 1000개를 발생시켜라.
data2 = data1 + (10*randn(1000) + 50)
print('data1 : mean = %.3f stdv = %.3f' % (mean(data1), std(data1)))
print('data2 : mean = %.3f stdv = %.3f' % (mean(data2), std(data2)))
pyplot.scatter(data1, data2)
pyplot.show() # 출력하라
# -
import matplotlib.pyplot as pit
print(pit.style.available)
# +
pit.style.use(['dark_background'])
xs = np.random.normal(0,3,(100,3)) # 평균이 0, 표준편차가 3인 100행 3열을 생성
pit.figure(figsize = (12,4)) # 모양을 결정
for i in range(0,3):
x = xs[:, i]
pit.plot(range(0, len(x)), x, linewidth=1, linestyle='--', label='x_{}'.format(i))
pit.legend() # 범례
pit.grid(False) # 격자선
pit.show()
# +
fig = pit.figure()
ax1 = fig.add_subplot(2,1,1)
ax2 = fig.add_subplot(2,1,2)
x = range(0,100)
y = [v*v for v in x]
ax1.plot(x,y)
ax2.bar(x,y) # 막대그래프
pit.title("Plot")
pit.show()
# +
x1 = np.linspace(0.0,5.0)
x2 = np.linspace(0.0,2.0)
pit.plot(np.exp(x1), "yo-") # 지수함수의 그래프
pit.show()
# +
y1 = np.cos(2 * np.pi * x1) * np.exp(-x1) # -x1으로 해놔서 줄어듦.
y2 = np.cos(2 * np.pi * x2)
ax1 = pit.subplot(2,1,1)
pit.plot(x1,y1, 'yo-') # Yellow(y), point marking(o), linetype(-)
pit.title("subplots")
print(ax1)
ax2 = pit.subplot(2,1,2)
pit.plot(x2, y2, 'm^-')
pit.xlabel('time(s)')
print(ax2)
pit.show()
# +
fig = pit.figure(figsize=(20,10))
ax1 = fig.add_subplot(121)
ax2 = fig.add_subplot(122)
# 이산형 막대그래프 bar, 연속 그래프 histogram
ax1.bar([1,2,3],[3,4,5], color = 'y')
ax2.barh([0.5,1,2.5],[0,1,2]) # v : vertical 수직으로, h : horizontal 수평으로
ax1.axvline(0,65)
ax2.axvline(0,45)
pit.tight_layout() # 여백을 줄여서 표현
pit.show()
# +
# boxplot
# IQR(Inter Quantile Range = 3 사분위수 - 1 사분위수)
# IQR * +-1.5 : 상하한선
pit.style.use(['classic'])
np.random.seed(19680801) # 시작점이 19680801
spread = np.random.rand(50) * 100 # 분포
center = np.ones(25) * 50 # 중심
filter_high = np.random.rand(10) * 100 + 100 # 상한선을 넘는 데이터
filter_low = np.random.rand(10) * -100
data = np.concatenate((spread, center, filter_high, filter_low)) # 데이터를 병합
fig1, ax1 = pit.subplots() # 형태를 제어, ax1은 '도화지'임.
ax1.set_title('boxplot')
ax1.boxplot(data)
# -
fig2, ax2 = pit.subplots()
ax2.set_title("Notched boxes") # 홈이 95% 신뢰구간
ax2.boxplot(data, notch = True)
# vertical : 수직의 , horizontal : 수평의
red_square = dict(markerfacecolor='r', marker='s') # s = square
fig5, ax5 = pit.subplots()
ax5.set_title("Horizontal")
ax5.boxplot(data, vert=False, flierprops=red_square) # 이상치 표현을 제어
# +
#CNN : C Neural Network
# +
# meshgrid = 그물망 격자
# 그래프에 수식을 표현할 땐 : latex 문법 사용 -> $ ---- $ : 달러 표시 사이에 수식 표현
points = np.arange(-5,5,0.01) # 1000개 생성
xs, ys = np.meshgrid(points, points) # np.meshgrid(1000, 1000) :총 100만개의 정점이 생김
z = np.sqrt(xs**2 + ys**2) # center로부터의 거리값 계산
# 팔레트 :
pit.imshow(z, cmap= pit.cm.rainbow) # 데이터(숫자 값)를 이미지(사각형)로 계산 / imshow : 값을 컬러값으로 표현
pit.colorbar()
pit.title(" $\sqrt{x^2+y^2}$")
pit.show()
# +
#Axes3D = 3차원
from mpl_toolkits.mplot3d import Axes3D
fig = pit.figure()
ax = Axes3D(fig) # 3차원 출력 도화지 = ax
X = np.arange(-4,4,0.25)
Y = np.arange(-4,4,0.25)
X, Y = np.meshgrid(X,Y) # 좌표점으로 변환 됨. => -4, -3.75, -.3.5
R = np.sqrt(X**2 + Y**2) # 거리값
#Z = R
Z = np.sin(R)
ax.plot_surface(X,Y,Z, rstride=1, cstride=1, cmap='hot') # r = row, c = column => rstride = 행 간격, cstride = 열 간격
pit.show()
ax.plot_surface(X,Y,Z, rstride=1, cstride=1, cmap=pit.cm.rainbow)
pit.show()
# +
from mpl_toolkits.mplot3d import Axes3D
# 방정식
def f(x,y): return (1 - x/2 + x**5 + y**3) * np.exp(-x**2 -y**2) # -로 줬으니까 아래로 내려감.
fig = pit.figure()
ax = Axes3D(fig)
X = np.arange(-4, 4, 0.25)
Y = np.arange(-4, 4, 0.25)
X, Y = np.meshgrid(X, Y)
Z = np.sin(f(X,Y))
# row, column
ax.plot_surface(X,Y,Z, rstride=1, cstride=1, cmap='hot')
pit.show()
# -
pit.contourf(X,Y,f(X,Y), 8, alpha=.75, cmap='jet') # 등고선
pit.colorbar()
pit.show()
# +
a = [0,0,1,0,
1,0,1,0,
1,1,1,1,
0,0,1,0]
np1 = np.array(a)
print(np1)
pit.imshow(np1.reshape(4,4), cmap='Greys', interpolation='nearest')
# interpolation( = 보간법)이 필요한 이유 : 컬러값이 지정이 되지 않으면 보간해서 적용해야하기 때문.
pit.show()
# +
# %matplotlib inline
from matplotlib import font_manager, rc # font_manager : 폰트를 설정하는..
import matplotlib
import matplotlib.pyplot as plt
plt.style.use(["classic"])
font_path = "C:/Windows/Fonts/H2GTRM.TTF" # 윈도우 모든 폰트가 있는 곳
font_name = font_manager.FontProperties(fname=font_path).get_name() # 폰트이름 획득
# resource configuration 폰트 이름
matplotlib.rc('font', family = font_name)
plt.plot([1,2,3,4])
plt.xlabel("시간")
plt.ylabel("거리")
plt.show()
# +
import matplotlib.font_manager as fm
path = "C:\\Windows\\Fonts\\NanumBarunGothic.ttf"
fontprop = fm.FontProperties(fname = path, size = 18)
data = np.random.randint(-100, 100, 50).cumsum() # cumulative sum : 누적합계
data
plt.plot(range(50), data, 'r')
plt.title('가격변동 추이', fontproperties = fontprop)
plt.ylabel('가격', fontproperties = fontprop)
plt.show()
# +
from pylab import plt
plt.style.use('ggplot')
import matplotlib as mpl
mpl.rcParams['font.family'] = '바탕'
def f(x):
return np.sin(x) + 0.5 * x
x = np.linspace(-2 * np.pi, 2 * np.pi, 50)
plt.plot(x, f(x), 'b')
plt.grid(True)
plt.xlabel('x')
plt.ylabel('f(y)')
# +
# 최소제곱법을 이용해서 fitting 적합
# polyfit 다차 방정식으로 fitting(적합) : 계수를 찾아내는 것
reg = np.polyfit(x, f(x), deg=5) # degree = 5 : 5차 방정식으로 피팅해라 => 피팅을 하면 개수가 반환 됨.
ry = np.polyval(reg, x) # = np.polyval( np.polyfit(x, f(x), deg=5) , x)
plt.plot(x, f(x), 'b', label = 'f(x)') # 원본데이터
plt.plot(x, ry, 'r', label='regression') # 최소제곱법으로 피팅된 데이터
plt.legend(loc = 0 ) #범례
plt.grid(True) # 격자
plt.xlabel('x')
plt.ylabel('f(x)')
# +
# 지리정보시스템
import folium
#위경도 좌표
map_1 = folium.Map(location=[37.565711, 126.978090], zoom_start=16) #확대축소
#tiles='Stamen Terrain')
folium.Marker([37.565711, 126.978090], popup='서울시청'). add_to(map_1)
folium.Marker([37.565711, 126.978190], popup='서울시청 및', icon=folium.Icon(icon='cloud')).add_to(map_1)
map_1
# -
# # 숙제 : 서울 소재 10개 대학의 좌표를 출력하시오.
# +
import folium
#위경도 좌표
map_1 = folium.Map(location=[37.565711, 126.978090], zoom_start=13) #확대축소
#tiles='Stamen Terrain')
folium.Marker([37.459329, 126.953105], popup='서울대학교'). add_to(map_1)
folium.Marker([37.561860, 126.946828], popup='이화여자대학교').add_to(map_1)
folium.Marker([37.583927, 127.059013], popup='서울시립대학교').add_to(map_1)
folium.Marker([37.556407, 126.987478], popup='숭의여자대학교').add_to(map_1)
folium.Marker([37.480173, 126.996354], popup='백석예술대학교').add_to(map_1)
folium.Marker([37.564984, 126.962323], popup='경기대학교 서울캠퍼스').add_to(map_1)
folium.Marker([37.551586, 126.924985], popup='홍익대학교').add_to(map_1)
folium.Marker([37.565763, 126.938428], popup='연세대학교').add_to(map_1)
folium.Marker([37.589218, 127.032691], popup='고려대학교').add_to(map_1)
folium.Marker([37.542738, 127.076335], popup='건국대학교').add_to(map_1)
map_1
# -
# ## pandas : Series(1차원), Dataframe(2차원), Panel
# ## 판다스의 base = numpy => 넘파이 방식의 인덱싱이 가능
# ## 판다스는 넘파이에 dict를 더한 것 -> 중복 허용, 순서를 보장해서 데이터를 처리한다. : key indexing
import pandas as pd
data = {'a' : 0., 'b':1., 'c':2.} # dict
s = pd.Series(data) # 1차원, 키가 인덱스로 변함.
print(s['a']) # 키이 인덱싱
s = pd.Series(data, index=['b','c','d','a']) # 행 이름
print(s['a'])
print(s['d']) # 데이터가 없기 때문에 nan 출력됨
list(s)
# +
s = pd.Series([1,2,3,4,5], index = ['a','b','c','d','e'])
print("인덱스에 의한 출력 ", s[0]) # 순서에 의한 인덱스
print()
print(s['a']) # 키에 의한 인덱스
print()
print(s[:3]) # numpy에서 썼던 방식
print("음수",s[-3:])
s['a'] = 100
print(s['a'])
print()
s['f']=10
print(s['f'])
print()
print("filtering에 의한 출력 ", s[s>4]) # 인덱스가 boolean으로 변함(true/false)
print()
print(s)
print()
print(s*2)
# +
data = np.array(['a','b','c','d']) # ndarray
# range : 결과값이 리스트
# arange : 결과값이 ndarray
# RangeIndex 객체가 자동으로 생성됨 : 0 ~ 4까지 step 1씩 => 결과값 : (순서가 있는) index
s = pd.Series(data) # ndarray를 이용해 시리즈 초기화
print("시리즈 데이터", s.values) #값만
print("시리즈 인덱스", s.index) # RangeIndex(start=0, stop=4, step = 1)
print("시리즈 초기화", s)
print("시리즈 인덱스의 값", s.index.values)
print(s[0])
print(s.head()) # tail()
print(s.value_counts()) # 도수분포표 개수를 세겠다.
print(s.value_counts(normalize = True)) # 상대도수분포표 / 사이즈 1로!
print(s.describe()) # 파이썬 -> 문자열인 경우 object로 나온다.
print("데이터 타입은 : ", s.dtypes)
print("차원", s.ndim)
print("차수", s.shape)
# +
sdata = {'Ohio':35000, 'Texas':71000, 'Oregon':16000, 'Utah':5000}
obj3 = pd.Series(sdata)
print(obj3)
print('----------')
print(obj3.shape)
print('----------')
states = ['California','Ohio','Oregon','Texas']
obj4 = pd.Series(sdata, index = states)
print(obj4)
print('----------')
print("시리즈 인덱스", obj3.index) # 그냥 index
print('----------')
print("시리즈 인덱스", obj4.index) # 얘도 그냥 index, dtype = objecy = 문자열
print('----------')
print("null이 있는가? ", pd.isnull(obj4)) # 데이터가 null인가?
print('----------')
print("결측치가 있는가? ", pd.notnull(obj4)) # null이 아닐 때 True <-> isnull()
print('----------')
print("객체 출력")
print('----------')
print("obj3을 출력합니다. ", obj3)
print('----------')
print("obj4를 출력합니다. ", obj4)
print('----------')
# 짝이 맞지 않기 때문에 index가 다름
# 근데 연산이 되네? => 짝이 없는 건 nan으로 뜸
print("연산결과를 출력합니다.", obj3 + obj4)
# -
s = pd.Series(['A','B','Aaba','Baca', np.nan, 'CABA', 'cat'])
s.str.count('a') # s안에 a가 몇 개씩 있는지..! 대소문자 구분!
s = pd.Series([1,2,2.5,3,3.5,4,5]) # 데이터가 7개 -> 이렇게 부드러운 곡선이 나올 수가 없는데?
# -> 데이터가 없는 부분은 커널로 예측! (가우시안 분포)
s.plot.kde() # kernel density estimate
plt.show()
s.plot.kde(bw_method=0.3) #과적합 : bin width
plt.show()
import pandas as pd
area = pd.Series({'California': 423967, 'Texas': 695662,
'New York': 141297, 'Florida': 170312,
'Illinois': 149995})
pop = pd.Series({'California': 38332521, 'Texas': 26448193,
'New York': 19651127, 'Florida': 19552860,
'Illinois': 12882135})
data = pd.DataFrame({'area':area, 'pop':pop})
data
data['area'] #인덱싱 접근 방법
data.area
# +
# 객체 비교 : is None
data.area is data['area']
# 객체에서 변수를 실시간으로 추가하는 것이 가능
# list -> append()
data['density'] = data['pop'] / data['area']
data
# -
data.values # ndarray => numpy의 함수들로 처리하면 됨
data.T # 데이터 전치
data.values[0] # 행 값
data.iloc[:3,:2] # 행과 열로 입력 : interger location : 순서에 의해 인덱싱 할 때
data.loc[:'Illinois', :'pop'] # 키에 의해 인덱싱 할 때 -> loc
data.iloc[0,2] = 90
data
data['Florida':'Illinois']
# +
import pandas as pd
import numpy as np
d = {'Name':pd.Series(['김하나','이하나','삼하나','사하나','오하나','육하나','칠하나', '팔하나']), # 이름 8개
'Age':pd.Series([25,26,25,23,30,29,23]), # 나이 7개
'Rating':pd.Series([4.23,3.24,3.98,2.56,3.20,4.6,3.8])} #이것도 7개
# -
df = pd.DataFrame(d)
print(df)
print('축', df.axes)
print('데이터 타입', df.dtypes) # 객체마다의 데이터 타입을 보여줌
print('데이터 타입', df.get_dtype_counts) # unique ( 중복이 없으면 전체 출력 )
print('비었나?', df.empty) # 데이터가 비었나? -> 아뇽
print('차원',df.ndim) # 2차원
print('차수',df.shape) # 8행 3열
print('사이즈',df.size) # 8*3 = 24
print('값', df.values) # 값만 출력
print(df.head(2)) # 맨 앞 2개 출력
print(df.tail(2)) # 맨 뒤 2개 출력
print("합계", df.sum()) # 전체 데이터의 합
print('--------------')
print(df.mean()) # 숫자만 적용
print('--------------')
print(df.std()) # standard deviation 표준편차 -> 숫자만 적용
print('↓describe()--------------')
print(df.describe()) # 숫자만 적용
print('↓include--------------')
print('오브젝트', df.describe(include=['object'])) # 만약 문자 데이터도 describe하고 싶다? -> include
print('--------------')
print(df.describe(include='all')) # uniqu, top, freq
# +
data={'state': ['경기', '강원', '서울', '충북', '인천'],
'year':[2000,2001,2002,2001,2002],
'pop':[1.5,1.7,3.6,2.4,2.9]} # dict
frame2=pd.DataFrame(data, columns=['year', 'state', 'pop', 'debt'],
index=['one', 'two', 'three', 'four', 'five']) # dataframe
# +
# 1) state만 출력하시오
print(frame2['state'])
print(frame2.state)
# 2) debt 열을 추가하고 모든 값을 16.5로 입력하시오.
# data['debt'] = [16.5,16.5,16.5,16.5,16.5]
# print(data['debt'])
frame2['debt']= 16.5
print("debt 값 적용 후 ", frame2)
# 3) debt를 pd.Series([-1.2,-1.5,-1.7], index=['two','four','five'])로 수정하시오
frame2.debt = pd.Series([-1.2,-1.5,-1.7], index=['two','four','five'])
frame2
# 4) 파생변수 'estern'에 주소가 서울인가를 따져 서울은 True 아니면 False가 입력되게 하시오.
# 파생변수 = 새롭게 만들어진 변수
frame2['estern'] = frame2.state == '서울'
print(frame2.columns)
# 특정 변수를 지울 때 : del frame2['estern']
print(frame2["year"]["one"]) # 열이 먼저 옴
print(frame2.loc["one","year"])
frame2
# +
# 시간 함수
# 리눅스 => timestamp : 1970-01-01 이후로 경과된 mili초로 표현
# 내부적으로 데이터 저장시 timestamp
# pandas => nano 초까지 표현!
# 시간 인덱스 : 시계열분석 : DateTimeindex, Periodindex 주기값
# 파이썬 : time.time() : 2000-01-01부터 경과된 mili초로 표현
# datetime(날짜와 시간을 표현)
# -
import time
print(time.time()) # 말라초로 표현 : . 밑에가 밀리초
print(time.localtime()) # 년월일 시분초
yesterday = time.localtime(time.time() - 60*60*24) # 시간 연산
yesterday
print(yesterday)
time.strftime('%Y %m %d') # strftime(): 시간을 포맷에 맞게 문자열로 출력
# +
from datetime import date, time, datetime, timedelta # 날짜, 시간, 날짜와 시간, 시간차
now = datetime.now() # 년월일 시분초
print("현재 :", now)
print(now.year, now.month, now.day)
now.timestamp() # 밀리초로 표현 ( . 이하)
# -
now_str=now.strftime('%Y-%m-%d %H:%M:%S') # 년월일시분초 => 문자열
print(now_str)
datetime.strptime(now_str, '%Y-%m-%d %H:%M:%S') #2019-11-01 11:49:09 <-이런 걸 짜 형식으로 변환
delta = datetime(2020,1,7) - datetime(1996,4,4)
print("시간 차는 = ", delta) # 8678 days 차이가 난다..!
start = datetime(2020,1,7)
print(start + timedelta(12)) # 기본이 날짜. timedelta()가 뭘까? - 찾아보기
start + timedelta(hours = -5)
import pandas as pd
print(pd.datetime.now())
print(pd.Timestamp('2017-03-01'))
print(pd.Timestamp(1587687255, unit='s'))
# +
# DatetimeIndex 생성 : 행 : index, 열키 : columns
print(pd.date_range("11:00","13:30",freq="30min")) # 11시부터 13시 30분까지 30분 간격으로 출력하라
print()
print("시간으로", pd.date_range("11:00","13:30", freq = "30min").time)
print()
print(pd.date_range("11:00","13:30",freq="H")) # freq = "H" : 간격 = 시간 단위로!
# +
# DatetimeIndex
print(pd.to_datetime(pd.Series(['Jul 31. 2009','2010-01-10', None]))) # 지정된 시간을 인덱스로 만듦.
print()
print(pd.to_datetime(['2005/11/23', '2010.12.31', None])) # 매개변수에 들어오는 걸 데이터로 만듦.
# -
print(pd.date_range('1/1/2017', periods=5)) # periods = 5 -> 1일부터 5일동안! : 5개 출력!!
# 일 간격 : 기본주기는 날짜
print(pd.date_range('1/1/2017', periods=5, freq = 'M')) # 간격은 월 단위, 5개 출력! -> 1,2,3,4,5월
# +
ts = pd.Series(np.random.randn(1000), index = pd.date_range('1/1/2000', periods=1000))
ts = ts.cumsum()
ts.plot()
df = pd.DataFrame(np.random.randn(1000,4), index=ts.index, columns=list('ABCD')) # 1000 by 4 : 4종목 데이터 생성 -> A,B,C,D
dt = df.cumsum()
df.plot()
df.plot.bar()
# +
# 문제 다음 데이터를 데이터프레임에 저장하세요.
# 지역 2018 2017 2016 2015 2017-2018 증가율
# 서울 수도권 9904312 9631482 9762546 9853972 0.0283
# 부산 경상권 3448737 3393191 3512547 3655437 0.0163
# 인천 수도권 2890451 2632035 2517680 2466338 0.0982
# 대구 경상권 2466052 2431774 2456016 2473990 0.0141
# +
# columns = ["지역","2015","2016","2017","2018","2015-2018 증가율"]
# index = ["서울","부산","인천","대구"]
data = {'지역':['서울','부산','인천','대구'],
'2015':[9853972,3655437,2466338,2473990],
'2016':[9762546,3512547,2517680,2456016],
'2017':[9631482,3393191,2632035,2431774],
'2018':[9904312,3448737,2890451,2466052],
'2017-2018 증가율':[0.0283,0.0163,0.0982,0.0141]
}
rate = pd.DataFrame(data, columns=['지역','2015','2016','2017','2018','2017-2018 증가율'],
index = ['서울','부산','인천','대구'])
rate
# -
# 문제 : "2017-2018 증가율"을 %로 변경하시오.
rate['2017-2018 증가율'] = rate['2017-2018 증가율'] * 100
rate
# +
# 문제 : 2015 ~ 2017의 증가율을 구해서 변수를 추가하시오. ( 열이름 : "2015-2017 증가율")
# -
rate['2015-2017 증가율'] = (rate['2017'] - rate['2015']) / rate['2015'] * 100
rate
| 17,700 |
/Multiclass_standard_approach.ipynb | 59f7211f5e0af36babdabbe40d7650576a420922 | [] | no_license | z3193631/NLP_Demo | https://github.com/z3193631/NLP_Demo | 0 | 0 | null | null | null | null | Jupyter Notebook | false | false | .py | 27,588 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Python 3
# name: python3
# ---
# + [markdown] id="view-in-github" colab_type="text"
# <a href="https://colab.research.google.com/github/z3193631/NLP_Demo/blob/master/Multiclass_standard_approach.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# + [markdown] id="3zvajfrh3k2O" colab_type="text"
# # Bag of Words Approach and Logistic Regression for NLP
# + id="Xf-ywkZBq-SE" colab_type="code" colab={}
import pandas as pd, numpy as np
from sklearn.linear_model import LogisticRegression
from sklearn.feature_extraction.text import CountVectorizer, TfidfVectorizer
from sklearn.metrics import roc_auc_score
# + id="ywg0dDGku2t9" colab_type="code" colab={}
train = pd.read_csv('train_sample.csv')
vals = pd.read_csv('val_sample.csv')
test = pd.read_csv('test.csv.zip')
test_label = pd.read_csv('test_labels.csv.zip')
# + id="Mwsg6r2w2a5L" colab_type="code" colab={}
# from google.colab import drive
# drive.mount('/content/drive')
# + id="5nZmI8I_w6gC" colab_type="code" colab={}
join_test = pd.merge(test,test_label, how='inner')
final_test = join_test[join_test.ne(-1).all(axis=1)]
# + id="9L-m37EtxtnT" colab_type="code" outputId="17defcc2-5471-4590-96f6-b777c6ee49e6" colab={"base_uri": "https://localhost:8080/", "height": 34}
lens = train.comment_text.str.len()
lens.mean(), lens.std(), lens.max()
# + id="7mjhSJ-zyF0y" colab_type="code" outputId="3be2fa62-0aec-40cd-8978-1dda107e4713" colab={"base_uri": "https://localhost:8080/", "height": 286}
lens.hist()
# + id="Y25pRBxKyWIp" colab_type="code" outputId="2d991806-2c36-4227-c1b6-fc913179bc04" colab={"base_uri": "https://localhost:8080/", "height": 297}
label_cols = ['toxic', 'severe_toxic', 'obscene', 'threat', 'insult', 'identity_hate']
train['none'] = 1-train[label_cols].max(axis=1)
train.describe()
# + id="FrX22nVH0uWq" colab_type="code" outputId="fdb263b0-6170-4425-a71f-cbae99ea719a" colab={"base_uri": "https://localhost:8080/", "height": 34}
len(train),len(test)
# + id="nIL9CmiZ0w39" colab_type="code" colab={}
#making sure column is not empty
COMMENT = 'comment_text'
train[COMMENT].fillna("unknown", inplace=True)
test[COMMENT].fillna("unknown", inplace=True)
# + [markdown] id="F75WW_cN3blH" colab_type="text"
# ## Model Building
# + id="w0qE_x863UYj" colab_type="code" colab={}
##create ngrams
# import re, string
# re_tok = re.compile(f'([{string.punctuation}“”¨«»®´·º½¾¿¡§£₤‘’])')
# def tokenize(s): return re_tok.sub(r' \1 ', s).split()
# + id="DyBo2dP15NOW" colab_type="code" colab={}
# #This creates a sparse matrix with only a small number of non-zero elements (stored elements in the representation below).
# n = train.shape[0]
# vec = TfidfVectorizer(ngram_range=(1,2), tokenizer=tokenize,
# min_df=3, max_df=0.9, strip_accents='unicode', use_idf=1,
# smooth_idf=1, sublinear_tf=1 )
# trn_term_doc = vec.fit_transform(train[COMMENT])
# test_term_doc = vec.transform(test[COMMENT])
# trn_term_doc, test_term_doc
# + id="VwYa2hXC5fJN" colab_type="code" colab={}
# #naive bayes formula
# def pr(y_i, y):
# p = x[y==y_i].sum(0)
# return (p+1) / ((y==y_i).sum()+1)
# + id="jYzoxm1h6Nw2" colab_type="code" colab={}
# x = trn_term_doc
# test_x = test_term_doc
# + id="btNC98SQ6y3k" colab_type="code" colab={}
# #abstraction for the model steps
# def get_mdl(y):
# y = y.values
# r = np.log(pr(1,y) / pr(0,y))
# m = LogisticRegression(C=4, dual=True)
# x_nb = x.multiply(r)
# return m.fit(x_nb, y), r
# #fit one model for each dependent variable
# preds = np.zeros((len(test), len(label_cols)))
# for i, j in enumerate(label_cols):
# print('fit', j)
# m,r = get_mdl(train[j])
# preds[:,i] = m.predict_proba(test_x.multiply(r))[:,1]
# pred_test = m.predict_proba(test_x.multiply(r))[:,1]
# print('ROC AUC:', roc_auc_score(test_label[j], pred_test))
# loss.append(roc_auc_score(test_label[j], pred_test))
# + id="DpxufS2Y9maE" colab_type="code" colab={}
# df = pd.concat([train['comment_text'], vals['comment_text']], axis=0)
# df = df.fillna("unknown")
# nrow_train = train.shape[0]
# vectorizer = TfidfVectorizer(stop_words='english', max_features=50000)
# X = vectorizer.fit_transform(df)
# col = ['toxic', 'severe_toxic', 'obscene', 'threat', 'insult', 'identity_hate']
# preds = np.zeros((train.shape[0], len(col)))
# + id="-YMU6lihJeBN" colab_type="code" colab={}
df = train['comment_text']
df = df.fillna("unknown")
nrow_train = train.shape[0]
vectorizer = TfidfVectorizer(stop_words='english', max_features=50000)
X = vectorizer.fit_transform(df)
col = ['toxic', 'severe_toxic', 'obscene', 'threat', 'insult', 'identity_hate']
preds = np.zeros((train.shape[0], len(col)))
# + id="B_3ahKl5JfMx" colab_type="code" outputId="984f81a7-d353-4d22-dd03-4ff029e1c77f" colab={"base_uri": "https://localhost:8080/", "height": 442}
loss = []
Y = vectorizer.transform(final_test['comment_text']) #for test dataset validation
for i, j in enumerate(col):
print('===Fit '+j)
model = LogisticRegression(penalty='l2')
model.fit(X, train[j])
preds[:,i] = model.predict_proba(X)[:,1]
pred_train = model.predict_proba(X)[:,1]
pred_test = model.predict_proba(Y)[:,1]
print('ROC AUC:', roc_auc_score(final_test[j], pred_test))
loss.append(roc_auc_score(final_test[j], pred_test))
print('mean column-wise ROC AUC:', np.mean(loss))
# + id="JKMMEal8BZ_z" colab_type="code" colab={}
| 5,686 |
/in_class/week_12/in_class_demos_week12.ipynb | d12539e4e86e906dff44f8df71acfdf458a72cce | [
"MIT"
] | permissive | zachary-trozenski/ate252_ccac | https://github.com/zachary-trozenski/ate252_ccac | 0 | 0 | null | null | null | null | Jupyter Notebook | false | false | .py | 23,554 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# ## pandas-profiling Epilepsy QLD Donor Information
# !pip install pandas-profiling
# ### Import libraries
from IPython.core.debugger import set_trace
import pandas as pd
import pandas_profiling
# ### Load and prepare dataset
df=pd.read_csv("donor_information.csv", parse_dates=['Aquisition Date', 'Dob'], encoding='UTF-8')
# ### Inline report without saving object
pandas_profiling.ProfileReport(df)
# ### Save report to file
pfr = pandas_profiling.ProfileReport(df)
pfr.to_file("donor_information.html")
#### Print existing ProfileReport object inline
pfr
'atr')
ann_list = np.array(ann.sample)
return np.array(ann_list)
# -
ss = pd.read_csv("./Signal_Data/signals_anns.csv",sep=",")
ss = np.array(ss)
# +
ann_list =[]
ann_locations = []
ann_labels = []
str1 = "./Data_thesis/MIT/"
MIT_str = ["100","101","102","103","104","105","106","107","108","109","111","112","113","114","115","116","117","118","119",
"121","122","123","124","200","201","202","203","205","207","208","209","210","212","213","214","215","217",
"219","220","221","222","223","228","230","231","232","233","234"]
for i in MIT_str:
ann_list.append(annotation_symbols(str1,i))
ann_locations.append(annotation_locations(str1,i))
ann_labels.append(annotation_labels(str1,i))
str2 = "./Data_thesis/CUDB/"
cudb_str = ["cu01","cu02","cu03","cu04","cu05","cu06","cu07","cu08","cu09","cu10","cu11","cu12","cu13","cu14","cu15","cu16",
"cu17","cu18","cu19","cu20","cu21","cu22","cu23","cu24","cu25","cu26","cu27","cu28","cu29","cu30","cu31","cu32",
"cu33","cu34","cu35"]
for i in cudb_str:
ann_list.append(annotation_symbols(str2,i))
ann_locations.append(annotation_locations(str2,i))
ann_labels.append(annotation_labels(str2,i))
str3 = "./Data_thesis/VFDB/"
vfdb_str = ["418","419","420","421","422","423","424","425","426","427","428","429","430","602","605","607",
"609","610","611","612","614","615"]
for i in vfdb_str:
ann_list.append(annotation_symbols(str3,i))
ann_locations.append(annotation_locations(str3,i))
ann_labels.append(annotation_labels(str3,i))
# -
ann_list=np.array(ann_list)
ann_locations = np.array(ann_locations)
ann_labels = np.array(ann_labels)
wfdb.io.show_ann_labels()
wfdb.io.show_ann_classes()
def fib_location_VF(lis,loc,lis2):
listlist = []
listmist = []
for i in range(len(lis)):
for j in range(len(lis[i])):
if lis[i][j] == '(VF' or lis[i][j]=='(VFL' or lis[i][j] == '(VFIB':
for k in range(len(lis[i][j+1:-1])):
if lis[i][j+k+1] != '':
lastone = loc[i][j+k]
duration = lastone - loc[i][j-1]
break
if i <=47:
listlist.append(((i)*len(ss[i]))+int(loc[i][j]/len(ss[i][0])))
listmist.append(((i)*len(ss[i]))+int(loc[i][j+k]/len(ss[i][0])))
elif 47 < i < 83:
listlist.append((((i-47)*len(ss[i]))+(196*47))+int(loc[i][j]/len(ss[i][0])))
listmist.append((((i-47)*len(ss[i]))+(196*47))+int(loc[i][j+k]/len(ss[i][0])))
elif i >= 83:
listlist.append((((i-82)*len(ss[i]))+(196*47)+(38*34))+int(loc[i][j]/len(ss[i][0])))
listmist.append((((i-82)*len(ss[i]))+(196*47)+(38*34))+int(loc[i][j+k]/len(ss[i][0])))
for u in range(len(lis2)):
for v in range(len(lis2[u])):
if lis2[u][v] == '[':
for qq in range(len(lis2[u][v+1:-1])):
if lis2[u][v+qq] == ']':
lastone = loc[u][v+qq]
duration = lastone - loc[u][v-1]
break
else:
break
if u <=47:
listlist.append(((u)*len(ss[u]))+int(loc[u][v]/len(ss[u][0])))
listmist.append(((u)*len(ss[u]))+int(loc[u][v+qq]/len(ss[u][0])))
elif 47 < u < 83:
listlist.append((((u-47)*len(ss[u]))+(196*47))+int(loc[u][v]/len(ss[u][0])))
listmist.append((((u-47)*len(ss[u]))+(196*47))+int(loc[u][v+qq]/len(ss[u][0])))
elif u >= 83:
listlist.append((((u-82)*len(ss[u]))+(196*47)+(38*34))+int(loc[u][v]/len(ss[u][0])))
listmist.append((((u-82)*len(ss[u]))+(196*47)+(38*34))+int(loc[u][v+qq]/len(ss[u][0])))
elif lis2[u][v] == '!':
for nn in range(len(lis2[u][v+1:-1])):
if lis2[u][v+nn+1] != '!' and lis2[u][v+nn+1] != ['"', '+', '/', 'A', 'E', 'F', 'J', 'L', 'N', 'Q', 'R', 'S','V', '[',
']', 'a', 'e', 'f', 'j', 'x', '|', '~']:
lastone = loc[u][v+nn]
duration = lastone - loc[u][v-1]
break
if u <=47:
listlist.append(((u)*len(ss[u]))+int(loc[u][v]/len(ss[u][0])))
listmist.append(((u)*len(ss[u]))+int(loc[u][v+nn]/len(ss[u][0])))
elif 47 < u < 83:
listlist.append((((u-47)*len(ss[u]))+(196*47))+int(loc[u][v]/len(ss[u][0])))
listmist.append((((u-47)*len(ss[u]))+(196*47))+int(loc[u][v+nn]/len(ss[u][0])))
elif u >= 83:
listlist.append((((u-82)*len(ss[u]))+(196*47)+(38*34))+int(loc[u][v]/len(ss[u][0])))
listmist.append((((u-82)*len(ss[u]))+(196*47)+(38*34))+int(loc[u][v+nn]/len(ss[u][0])))
return np.array(listlist),np.array(listmist)
sm = pd.read_csv("./Signal_Data/signals.csv",sep = ",")
del sm['Unnamed: 0']
np.array(sm)[0
VF_anns = fib_location_VF(ann_list,ann_locations,ann_labels)
# 1. 3 Premature ventricular contractions = VT
# 2. ! = VFL
# 3. Square brackets (open and closed) = VF
# 4. R on T premature contracitons = VT
def fib_location_VT(lis,loc,lis2):
listlist = []
listmist = []
for i in range(len(lis)):
for j in range(len(lis[i])):
if lis[i][j] == '(SVTA' or lis[i][j]=='(VT':
for k in range(len(ann_list[i][j+1:-1])):
if ann_list[i][j+k+1] != '':
lastone = loc[i][j+k]
duration = lastone - loc[i][j-1]
break
if i <=47:
listlist.append(((i)*len(ss[i]))+int(loc[i][j]/len(ss[i][0])))
listmist.append(((i)*len(ss[i]))+int(loc[i][j+k]/len(ss[i][0])))
elif 47 < i < 83:
listlist.append((((i-47)*len(ss[i]))+(196*47))+int(loc[i][j]/len(ss[i][0])))
listmist.append((((i-47)*len(ss[i]))+(196*47))+int(loc[i][j+k]/len(ss[i][0])))
elif i >= 83:
listlist.append((((i-82)*len(ss[i]))+(196*47)+(38*34))+int(loc[i][j]/len(ss[i][0])))
listmist.append((((i-82)*len(ss[i]))+(196*47)+(38*34))+int(loc[i][j+k]/len(ss[i][0])))
for u in range(len(lis2)):
cc = 0
for v in range(len(lis2[u])):
if lis2[u][v] == 'V':
cc +=1
else:
if v == len(lis2[u])-1:
cc = 0
elif lis2[u][v+1] == ['!', '"', '+', '/', 'A', 'E', 'F', 'J', 'L', 'N', 'Q', 'R', 'S','V', '[', ']', 'a', 'e', 'f', 'j', 'x', '|', '~']:#and lis2[u][v+2]=='+':
cc +=1
if cc ==4:
for qq in range(len(lis2[u][v-4:-1])):
if lis2[u][v+qq] != 'V':
lastone = loc[u][v+qq]
duration = lastone - loc[u][v-1]
break
else:
break
if u <=47:
listlist.append(((u)*len(ss[u]))+int(loc[u][v-3]/len(ss[u][0])))
listmist.append(((u)*len(ss[u]))+int(loc[u][v+qq-1]/len(ss[u][0])))
elif 47 < u < 83:
listlist.append((((u-47)*len(ss[u]))+(196*47))+int(loc[u][v-3]/len(ss[u][0])))
listmist.append((((u-47)*len(ss[u]))+(196*47))+int(loc[u][v+qq-1]/len(ss[u][0])))
elif u >= 83:
listlist.append((((u-82)*len(ss[u]))+(196*47)+(38*34))+int(loc[u][v-3]/len(ss[u][0])))
listmist.append((((u-82)*len(ss[u]))+(196*47)+(38*34))+int(loc[u][v+qq-1]/len(ss[u][0])))
return np.array(listlist),np.array(listmist)
VT_anns = fib_location_VT(ann_list,ann_locations,ann_labels)
def label_creator(anns):
labels = np.zeros(14236)
labels = np.array(labels)
i = 0
counter = 0
while 3 > 2:
for l in range(anns[0][i],anns[1][i]+1):
labels[l] = 1
counter+=1
i+=1
if i == len(anns[0]):
break
return pd.DataFrame(labels)
VF_labels = label_creator(VF_anns)
counter1 = 0
for i in np.array(VF_labels):
if i ==1:
counter1+=1
counter1
VT_labels = label_creator(VT_anns)
counter2 = 0
for i in np.array(VT_labels):
if i == 1:
counter2 +=1
counter2
counter1+counter2
v = []
for i in range(len(ann_list)):
for j in range(len(ann_list[i])):
v.append(ann_list[i][j])
np.unique(np.array(v))
v = []
for i in range(len(ann_labels)):
for j in range(len(ann_labels[i])):
v.append(ann_labels[i][j])
np.unique(np.array(v))
VT = pd.DataFrame(VT_labels)
VT.columns = ["VT"]
VF = pd.DataFrame(VF_labels)
VF.columns = ["VF"]
VF.to_csv("./Parameter_Data/VF_final.csv",sep=',')
VT.to_csv("./Parameter_Data/VT_final.csv",sep=',')
['str'] = "GeeksforGeeks"
d['x'] = 20
return d
# Driver code to test above method
d = fun()
print(d)
# + id="eAdD99Z1DDvm" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 34} outputId="8f38bbbb-f626-469b-f215-1de4bd90695f"
from functools import partial
def f(a,b,c,x):
return 1000*a + 100*b + 10*c +x
p = partial(f,4,5,6)
print(p(7))
# + id="L7oi0d_8GPzj" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 34} outputId="61845ff6-0fb2-4c53-bc10-8f0cbd4473cf"
from functools import *
# A normal function
def add(a, b, c):
return 100 * a + 10 * b + c
# A partial function with b = 1 and c = 2
add_part = partial(add, c = 2, b = 1)
# Calling partial function
print(add_part(3))
# + id="98sK-so6Ggf2" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 52} outputId="6b7fb9d3-675d-4f37-ce84-1c655814f496"
def shout(text):
print(text.upper())
def whisper(text):
print(text.lower())
def greetings(func, func2):
func("Hello there i am here !")
func2("Second fucntion using for whisper")
greetings(shout,whisper)
# + id="0QwHr68dHsX3" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 121} outputId="64427eb5-fe4f-40fa-f06a-89b6c5b48de7"
s = 'hekko ther i am goin fine'
for i in s.split(" "):
for j in range(len(i)):
if j==0:
a = i[j].upper()
m = a+i[1:]
print(m)
# + id="Ugs7W0QYIM-U" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 104} outputId="ce7ce523-5e3a-4094-8d0f-f31c7fadc052"
s = 'Heiko'
for i in s.split(" "):
for j in range(len(i)):
if i[j].isupper():
a = i[j].lower()
m = i[:j]+a+i[j+1:]
print(m)
if i[j].islower():
a = i[j].upper()
m = i[:j]+a+i[j+1:]
print(m)
# + id="ptOVqbPlJ0R0" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 34} outputId="8f9c220c-2b91-4a6b-9285-63456119548e"
def create(x):
def adder(y):
return x+y
return adder
add = create(20)
add(4)
# + id="IYmbOh4_Ov_R" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 69} outputId="10c77d5c-4b1b-482c-a8e3-bf0f02d1d065"
#1. trunc() :- This function is used to eliminate all decimal part
# of the floating point number and return the integer without the
# decimal part.
import math
a = 5.67
b = 4.4
print(math.trunc(a))
print(math.ceil(a))
print(math.floor(a))
# + id="NSDYq76cPH6A" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 34} outputId="f70ed723-9209-45fb-f4c9-e79f5e64a69c"
a = 3.53848752
round(a,4)
# + id="lID41Nl-QCD9" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 86} outputId="1163a54b-e14d-4fe2-d672-f09b01d730c8"
# Python program to illustrate
# *args with first extra argument
def myFun(arg1, *argv):
print ("First argument :", arg1)
for arg in argv:
print("Next argument through *argv :", arg)
myFun('Hello', 'Welcome', 'to', 'GeeksforGeeks')
# + id="qMDAziCLQ-XL" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 86} outputId="60ef48bd-dc03-4164-b58e-dfe8fc52783e"
def myFun(arg1, *argv):
print ("First argument :", arg1)
for arg in argv:
print("Next argument through *argv :", arg)
t = ( 'Welcome', 'to', 'GeeksforGeeks')
myFun('Hello',*t)
# + id="DyosdtCwRD-7" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 52} outputId="39590523-2463-48e1-f4f5-1bc332780b8e"
def myFun(*args,**kwargs):
print("args: ", args)
print("kwargs: ", kwargs)
# Now we can use both *args ,**kwargs to pass arguments to this function :
myFun('geeks','for','geeks',first="Geeks",mid="for",last="Geeks")
# + id="vaWIxkg1Rhlb" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 121} outputId="a3feecf0-de45-4a75-95da-ff6c73ac5a13"
def myFun(arg1, arg2, arg3):
print("arg1:", arg1)
print("arg2:", arg2)
print("arg3:", arg3)
# Now we can use *args or **kwargs to
# pass arguments to this function :
args = ("Geeks", "for", "Geeks")
myFun(*args)
kwargs = {"arg1" : "Geeks", "arg2" : "for", "arg3" : "Geeks"}
myFun(**kwargs)
# + id="anclYcuoRtX3" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 34} outputId="24697a2d-bcfa-47ed-bbb7-0deb2f883d69"
def outer(text):
text = text
def inner():
print(text)
inner()
if __name__ == "__main__":
outer("hello")
# + id="ZMcqJ-QwUF_b" colab_type="code" colab={}
def outer(text):
text = text
def inner():
print(text)
return inner
outerf = outer("HEY")
outerf()
# + id="7Rk6tbJPVTYJ" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 86} outputId="a8d499a4-a8c9-49d7-c48e-a7340b53372b"
# Python program to illustrate
# closures
import logging
logging.basicConfig(filename='example.log', level=logging.INFO)
def logger(func):
def log_func(*args):
logging.info(
'Running "{}" with arguments {}'.format(func.__name__, args))
print(func(*args))
# Necessary for closure to work (returning WITHOUT parenthesis)
return log_func
def add(x, y):
return x+y
def sub(x, y):
return x-y
add_logger = logger(add)
sub_logger = logger(sub)
add_logger(3, 3)
add_logger(4, 5)
sub_logger(10, 5)
sub_logger(20, 10)
# + id="t2-CUIwQXU4e" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 34} outputId="e8c454da-f6eb-428c-e449-723d83c8bf3e"
def msgwelcome(msg):
def addwelcome():
return "welcome to"
return addwelcome()+msg
msgwelcome(" Website")
# + id="67VsujaEYUln" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 34} outputId="a4d747cb-9c18-476f-a494-330e24b37eac"
# Adds a welcome message to the string
# returned by fun(). Takes fun() as
# parameter and returns welcome().
def decorate_message(fun):
# Nested function
def addWelcome(site_name):
return "Welcome to " + fun(site_name)
# Decorator returns a function
return addWelcome
@decorate_message
def site(site_name):
return site_name;
# Driver code
# This call is equivalent to call to
# decorate_message() with function
# site("GeeksforGeeks") as parameter
print(site("GeeksforGeeks"))
# + id="VYqIEMMQajPk" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 52} outputId="bc1a80fb-18c4-4c97-880d-f93b1ab68610"
# A Python example to demonstrate that
# decorators can be useful attach data
# A decorator function to attach
# data to func
def attach_data(func):
func.data = 3
return func
@attach_data
def add (x, y):
return x + y
# Driver code
# This call is equivalent to attach_data()
# with add() as parameter
print(add(2, 3))
print(add.data)
# + id="iORvL1L0aqPB" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 34} outputId="8e36f5e7-d6e9-4b98-a2a7-e2aa3f26dac1"
next()
# + id="FMKQs3MrjTGu" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 34} outputId="aa7b861b-a998-4a2f-8974-1070da774daa"
range(10)
# + id="eeKdXpB_ju5P" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 34} outputId="1c9254c5-654f-4d24-ec61-1393553a6479"
iter(range(10))
# + id="Zw-ZBnarjxrS" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 86} outputId="83d673f4-ecb0-4f09-ef0d-96b437480913"
demo = range(2,199,3)
print(demo)
print(demo.step)
print(demo.index(26))
print(demo.index(29))
# + id="Eb3I9y27kFWi" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 86} outputId="ffe2c68c-6812-4454-c947-e618f3c216fd"
def myfunction(text):
return text
'''returns the text given by the user '''
myfunction("nayem")
help(myfunction)
# + id="9HNUK1JvmTJ3" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 453} outputId="f1307d77-ad3e-4f05-8489-de715ffc7ed7"
class Helper:
def __init__(self):
'''The helper class is initialized'''
def print_help(self):
'''Returns the help description'''
print('helper description')
help(Helper)
help(Helper.print_help)
# + [markdown] id="7Hvgh4XVpZZe" colab_type="text"
# ## Exception handling in python
# + id="xj_-Xu9bmlgc" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 34} outputId="d43d3e7d-b4c8-4a69-c06c-8b5bfc0a08e6"
try :
n = 3
r = n/(n-3)
print(r)
except (ZeroDivisionError, NameError) as e:
print("Error occurred : ",e)
# + id="L9wRqLipobRX" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 52} outputId="e89b8a69-d58a-4e2b-b7dd-6b5b14d166dc"
def subtract(a,b):
try:
print(a/b)
except (ZeroDivisionError, TypeError, NameError) as e:
print("Error occured : ",e)
else:
print("subtraction done")
finally:
print("I am finally clause, always raised")
subtract(6,0)
# + id="ovC8eJSqobi_" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 34} outputId="fcd7d635-a666-4dcd-f896-c1aa76d7cb67"
try:
raise NameError("this is name error")
except NameError:
print("Name error is found")
# + id="KRO1c7Unobm-" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 341} outputId="c994a2c6-b293-44de-eb9c-ed1abd8b2dcf"
# Python code to illustrate
# clean up actions
def divide(x, y):
try:
# Floor Division : Gives only Fractional Part as Answer
result = x // y
except ZeroDivisionError:
print("Sorry ! You are dividing by zero ")
else:
print("Yeah ! Your answer is :", result)
finally:
print("I'm finally clause, always raised !! ")
# Look at parameters and note the working of Program
divide(3, "3")
# + id="EVAv2fj5obgu" colab_type="code" colab={}
# + id="J-CEcFTjobfL" colab_type="code" colab={}
# + id="a8usMh3sobdi" colab_type="code" colab={}
# + id="giIEusLKobbz" colab_type="code" colab={}
# + id="dE78wRH6obZp" colab_type="code" colab={}
# + id="rRsra_X_obYK" colab_type="code" colab={}
# + id="HpINGVQqobVd" colab_type="code" colab={}
# + id="0HzLWZOtobPS" colab_type="code" colab={}
# + id="8trcMRtwobNG" colab_type="code" colab={}
# + id="bqHw7-XuobLH" colab_type="code" colab={}
# + id="8r_qg4LpobI0" colab_type="code" colab={}
# + id="6xbPLvl9obHD" colab_type="code" colab={}
# + id="9Q7I1-bgobEl" colab_type="code" colab={}
# + id="Z6R8ffplobCT" colab_type="code" colab={}
# + [markdown] id="xD1mGUm_k6CG" colab_type="text"
# ### need to be checked
# coroutine- functions, memorization using decorators , decorators in python
| 20,961 |
/Ch09_RiskMeasures.ipynb | d1cf9bba2882eae9d7c8d874ef10568c0f9fc417 | [
"MIT"
] | permissive | mengphilshen/FinancialTheoryMSc | https://github.com/mengphilshen/FinancialTheoryMSc | 0 | 0 | null | null | null | null | Jupyter Notebook | false | false | .jl | 371,981 | # -*- coding: utf-8 -*-
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .jl
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Julia 1.0.3
# language: julia
# name: julia-1.0
# ---
# ## Load Packages
# +
using Dates, Distributions, DelimitedFiles
include("jlFiles/printmat.jl")
# +
using Plots
backend = "gr" #"gr" (default), "pyplot"
if backend == "pyplot"
pyplot(size=(600,400))
else
gr(size=(480,320))
default(fmt = :svg)
end
# -
# # VaR for a N(μ,σ²) Return
# $\textrm{VaR}_{95\%} = - (5^{th}$ percentile of the return distribution)
#
# With a $N(\mu,\sigma^2)$ distribution this gives
#
# $\textrm{VaR}_{95\%} = - (\mu-1.64\sigma)$
function ϕNS(x,μ=0,σ²=1) #pdf of N(μ,σ²), NS for "non-standard"
pdfx = pdf(Normal(μ,sqrt(σ²)),x) #the Distributions package wants σ in
return pdfx #Normal(μ,σ), not σ²
end
# +
μ = 8
σ = 16
q05 = μ - 1.64*σ
VaR95 = -(μ - 1.64*σ)
printlnPs("with μ=$μ and σ=$σ, the 5th quantile and VaR 95% are: ",[q05 VaR95])
# -
q05b = quantile(Normal(μ,σ),0.05) #exact calculation of the the 5th quantile
printlnPs("get an exact result by using the quantile() function: ",q05b)
# +
R = range(-60,stop=60,length=301)
pdfR = ϕNS.(R,μ,σ^2)
Rb = R[R .<= -VaR95]
p1 = plot(R,pdfR,color=:red,linewidth=2,label="pdf of N($μ,$(σ^2))")
plot!(Rb,ϕNS.(Rb,μ,σ^2),color=:red,linewidth=2,fill=(0,:blue),label="")
title!("Pdf and VaR")
xlabel!("return, %")
plot!([-VaR95],linetype=:vline,color=:blue,label="VaR (95%)")
# -
# # Loading Daily S&P 500 Data
# +
x = readdlm("Data/SP500RfPs.csv",',',skipstart=1)
SP = x[:,2] #S&P 500 level
R = (SP[2:end]./SP[1:end-1] .- 1) * 100 #returns, %
T = length(R)
dN = Date.(string.(x[2:end,1]),"d/m/y") #convert to Date, 2:end as for R
println("Number of days in the sample: $T")
# -
# # Backtesting VaR from N() on Data
# To backtest a VaR model, study the relative frequency of Loss > VaR.
#
# The code below does this for difference confidence levels (0.95,0.96,...) of the VaR.
# +
μ_emp = mean(R) #mean and std of data
σ_emp = std(R)
confLev = 0.95:0.005:0.995
L = length(confLev)
Loss = -R
VaR = fill(NaN,L)
BreakFreq = copy(VaR)
for i = 1:L #loop over different (1-confidence levels)
VaR[i] = -quantile(Normal(μ_emp,σ_emp),1-confLev[i])
BreakFreq[i] = mean(Loss .> VaR[i]) #frequency of breaking the VaR
end
println("conf level N()-based VaR actual coverage")
printmat([confLev VaR (1.0.-BreakFreq)],width=15)
# -
# The code below studes the relative frequency of Loss > VaR, but this time over a moving data window. This allows us to investigate if there are long periods of failure (in either direction) the VaR.
# +
VaR95 = -(μ_emp - 1.64*σ_emp)
BreakFreqT = fill(NaN,T) #vector, freq(Loss>VaR) on moving data window
for t = 101:T
BreakFreqT[t] = mean(Loss[t-100:t] .> VaR95)
end
xTicksLoc = Dates.value.([Date(1980);Date(1990);Date(2000);Date(2010)])
xTicksLab = ["1980";"1990";"2000";"2010"] #crude way of getting tick marks right
p1 = plot(dN,BreakFreqT*100,color=:blue,ylim=(-1,35),legend=false,
xticks=(xTicksLoc,xTicksLab))
title!("Frequency of Loss > VaR 95%")
ylabel!("%")
plot!([5],linetype=:hline,color=:black)
annotate!(Dates.value(Date(1980)),32,text("over the last 100 days",8,:left))
# -
# # A Simple Dynamic VaR with Time-Varying Volatility
# We first construct an simple estimate of $\sigma_t^2$ as a backward looking exponential moving average
#
# $\sigma_t^2 = \lambda \sigma_{t-1}^2 + (1-\lambda) (R_{t-1} -\mu_{t-1})^2$,
# where $\mu_{t}=\lambda \mu_{t-1} + (1-\lambda) R_{t-1}$
#
# Redo the VaR calculation using
#
# $\textrm{VaR}_{t} = - (\mu_t-1.64\sigma_t)$ and study if it has better properties than the static VaR
# +
λ = 0.94
s2T = fill(σ_emp^2,T) #vector, time-varying variance
μT = fill(μ_emp,T)
for t = 2:T
μT[t] = λ*μT[t-1] + (1-λ)*R[t-1]
s2T[t] = λ*s2T[t-1] + (1-λ)*(R[t-1]-μT[t-1])^2 #RiskMetrics approach
end
BreakFreq = fill(NaN,L)
for i = 1:L
local critval, VaR_i
critval = abs(quantile(Normal(0,1),1-confLev[i]))
VaR_i = -(μT .- critval*sqrt.(s2T))
BreakFreq[i] = mean(Loss .> VaR_i)
end
println("conf level, coverage")
printmat([confLev (1.0.-BreakFreq)])
# +
VaR95 = -(μT .- 1.64*sqrt.(s2T))
BreakFreqT = fill(NaN,T) #vector, freq(Loss>VaR) on moving data window
for t = 101:T
BreakFreqT[t] = mean(Loss[t-100:t] .> VaR95[t-100:t])
end
xTicksLoc = Dates.value.([Date(1980);Date(1990);Date(2000);Date(2010)])
xTicksLab = ["1980";"1990";"2000";"2010"] #crude way of getting tick marks right
p1 = plot(dN,BreakFreqT*100,color=:blue,ylim=(-1,35),legend=false,xticks=(xTicksLoc,xTicksLab))
title!("Frequency of Loss > VaR 95%")
ylabel!("%")
plot!([5],linetype=:hline,color=:black)
annotate!(Dates.value(Date(1980)),32,text("over the last 100 days",8,:left))
# -
# # Expected Shortfall
# Recall: $\text{ES}_{\alpha}=-\text{E}(R|R\leq-\text{VaR}_{\alpha})$
#
# For a normally distributed return $R\sim N(\mu,\sigma^{2})$ we have
#
# $\text{ES}_{95\%}=-\mu+\frac{\phi(-1.64)}{0.05}\sigma$
# +
μ = 8
σ = 16
ES95 = -(μ - ϕNS(1.64)/0.05*σ)
printlnPs("N() based ES 95% with μ=$μ and σ=$σ is: ",ES95)
# +
ESN = fill(NaN,L)
ES_emp = copy(ESN)
for i = 1:L
local critval, vv_i
critval = abs(quantile(Normal(0,1),1-confLev[i]))
ESN[i] = -(μ_emp .- ϕNS(critval)/(1-confLev[i])*σ_emp)
vv_i = Loss .> VaR[i]
ES_emp[i] = mean(Loss[vv_i]) #mean of obs when Loss > VaR
end
println("Conf level ES from N() ES (historical)")
printmat([confLev ESN ES_emp],width=12)
# -
| 5,945 |
/자료구조_이진 검색 트리.ipynb | c5505b9d003cdcb16ce9b51b0cac48e232451563 | [] | no_license | allan02/SAMSUNG_SDS_Algorithm | https://github.com/allan02/SAMSUNG_SDS_Algorithm | 0 | 0 | null | null | null | null | Jupyter Notebook | false | false | .py | 1,765 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# +
answer = []
import sys
sys.setrecursionlimit(20000)
def sol(array, left, right):
if left > right:
return
root = array[left]
temp = right + 1
for i in range(left + 1, right + 1):
if root < array[i]:
temp = i
break
sol(array, left + 1, temp - 1)
sol(array, temp, right)
answer.append(root)
values = []
while True:
try:
values.append(int(input()))
except:
break
sol(values, 0, len(values) - 1)
print('\n'.join(map(str,answer)))
| 831 |
/NLP_LSA_LDA_TSNE.ipynb | 6b0538447fd57f90e9db12204d7b297be1bf57a3 | [] | no_license | Amey-Mohite/NLP_assignments | https://github.com/Amey-Mohite/NLP_assignments | 7 | 0 | null | null | null | null | Jupyter Notebook | false | false | .py | 8,097,579 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
import nltk
import pandas as pd
from sklearn.feature_extraction.text import TfidfVectorizer
from sklearn.decomposition import TruncatedSVD
import nltk
from nltk.corpus import stopwords
from string import punctuation
nltk.download('stopwords')
nltk.download('punkt')
stop_words = stopwords.words('english')
from string import punctuation
custom = stop_words+list(punctuation)
from nltk.stem import WordNetLemmatizer
wordnet_lemmatizer = WordNetLemmatizer()
nltk.download('wordnet')
import warnings
warnings.filterwarnings("ignore")
def my_tokenizer(s):
s = s.lower()
tokens = nltk.tokenize.word_tokenize(s)
tokens = [t for t in tokens if len(t)>2] #remove words lesser than 2 in length
tokens = [wordnet_lemmatizer.lemmatize(t) for t in tokens] #lemmatize words
tokens = [t for t in tokens if t not in custom] #remove stopwords and punctuation
tokens = [t for t in tokens if not any(c.isdigit() for c in t)] # remove digits
return tokens
import os
os.getcwd()
df = pd.read_csv(r"G:\study\Natural Language processing\nlp_class\voted-kaggle-dataset.csv\voted-kaggle-dataset.csv")
df = df[["Description"]]
df = df.dropna()
df.columns = ["text"]
df.isnull().sum()
df
text = df["text"].tolist()
text = [my_tokenizer(s) for s in text]
text
import gensim
from gensim.corpora import dictionary
from gensim import corpora
from pprint import pprint
id2word = corpora.Dictionary(text)
print (id2word)
print(id2word.token2id)
mycorpus = [id2word.doc2bow(s) for s in text]
len(mycorpus)
mycorpus[0]
text[0]
lda_model = gensim.models.ldamodel.LdaModel(corpus=mycorpus,
id2word=id2word,
num_topics=4,
random_state=42,
update_every=1,
chunksize=100,
passes=10,
alpha='auto',
per_word_topics=True)
pprint(lda_model.print_topics())
doc_lda = lda_model[mycorpus]
print('\nPerplexity: ', lda_model.log_perplexity(mycorpus)) # a measure of how good the model is. lower the better.
from gensim.models import CoherenceModel
coherence_model_lda = CoherenceModel(model=lda_model, texts=text, dictionary=id2word, coherence='c_v')
coherence_lda = coherence_model_lda.get_coherence()
print('\nCoherence Score: ', coherence_lda)
import pyLDAvis
import pyLDAvis.gensim # don't skip this
import matplotlib.pyplot as plt
# %matplotlib inline
pyLDAvis.enable_notebook()
vis = pyLDAvis.gensim.prepare(lda_model, mycorpus, id2word)
vis
import os
# +
# mallet_path = "/Users/Arunabh/Desktop/mallet-2.0.8/bin/mallet"
# +
# ldamallet = gensim.models.wrappers.LdaMallet(mallet_path, corpus=mycorpus, num_topics=4, id2word=id2word)
# -
def compute_coherence_values(dictionary, corpus, texts, limit, start=2, step=3):
"""
Compute c_v coherence for various number of topics
Parameters:
----------
dictionary : Gensim dictionary
corpus : Gensim corpus
texts : List of input texts
limit : Max num of topics
Returns:
-------
model_list : List of LDA topic models
coherence_values : Coherence values corresponding to the LDA model with respective number of topics
"""
coherence_values = []
model_list = []
for num_topics in range(start, limit, step):
model = gensim.models.ldamodel.LdaModel(corpus=mycorpus, num_topics=num_topics, id2word=id2word)
model_list.append(model)
coherencemodel = CoherenceModel(model=model, texts=texts, dictionary=dictionary, coherence='c_v')
coherence_values.append(coherencemodel.get_coherence())
return model_list, coherence_values
model_list, coherence_values = compute_coherence_values(dictionary=id2word, corpus=mycorpus, texts=text, start=2, limit=40, step=6)
limit=40; start=2; step=6;
x = range(start, limit, step)
plt.plot(x, coherence_values)
plt.xlabel("Num Topics")
plt.ylabel("Coherence score")
plt.legend(("coherence_values"), loc='best')
plt.show()
for m, cv in zip(x, coherence_values):
print("Num Topics =", m, " has Coherence Value of", round(cv, 4))
optimal_model = model_list[2]
model_topics = optimal_model.show_topics(formatted=False)
pprint(optimal_model.print_topics(num_words=10))
# +
def format_topics_sentences(ldamodel=lda_model, corpus=mycorpus, texts=text):
# Init output
sent_topics_df = pd.DataFrame()
# Get main topic in each document
for i, row in enumerate(ldamodel[corpus]):
row = sorted(row, key=lambda x: (x[1]), reverse=True)
# Get the Dominant topic, Perc Contribution and Keywords for each document
for j, (topic_num, prop_topic) in enumerate(row):
if j == 0: # => dominant topic
wp = ldamodel.show_topic(topic_num)
topic_keywords = ", ".join([word for word, prop in wp])
sent_topics_df = sent_topics_df.append(pd.Series([int(topic_num), round(prop_topic,4), topic_keywords]), ignore_index=True)
else:
break
sent_topics_df.columns = ['Dominant_Topic', 'Perc_Contribution', 'Topic_Keywords']
# Add original text to the end of the output
contents = pd.Series(texts)
sent_topics_df = pd.concat([sent_topics_df, contents], axis=1)
return(sent_topics_df)
df_topic_sents_keywords = format_topics_sentences(ldamodel=optimal_model, corpus=mycorpus, texts=text)
# Format
df_dominant_topic = df_topic_sents_keywords.reset_index()
df_dominant_topic.columns = ['Document_No', 'Dominant_Topic', 'Topic_Perc_Contrib', 'Keywords', 'Text']
# Show
df_dominant_topic.head(10)
# +
def format_topics_sentences(ldamodel=None, corpus=mycorpus, texts=text):
# Init output
sent_topics_df = pd.DataFrame()
# Get main topic in each document
for i, row_list in enumerate(ldamodel[corpus]):
row = row_list[0] if ldamodel.per_word_topics else row_list
# print(row)
row = sorted(row, key=lambda x: (x[1]), reverse=True)
# Get the Dominant topic, Perc Contribution and Keywords for each document
for j, (topic_num, prop_topic) in enumerate(row):
if j == 0: # => dominant topic
wp = ldamodel.show_topic(topic_num)
topic_keywords = ", ".join([word for word, prop in wp])
sent_topics_df = sent_topics_df.append(pd.Series([int(topic_num), round(prop_topic,4), topic_keywords]), ignore_index=True)
else:
break
sent_topics_df.columns = ['Dominant_Topic', 'Perc_Contribution', 'Topic_Keywords']
# Add original text to the end of the output
contents = pd.Series(texts)
sent_topics_df = pd.concat([sent_topics_df, contents], axis=1)
return(sent_topics_df)
df_topic_sents_keywords = format_topics_sentences(ldamodel=lda_model, corpus=mycorpus, texts=text)
# Format
df_dominant_topic = df_topic_sents_keywords.reset_index()
df_dominant_topic.columns = ['Document_No', 'Dominant_Topic', 'Topic_Perc_Contrib', 'Keywords', 'Text']
df_dominant_topic.head(10)
# +
# Display setting to show more characters in column
pd.options.display.max_colwidth = 100
sent_topics_sorteddf_mallet = pd.DataFrame()
sent_topics_outdf_grpd = df_topic_sents_keywords.groupby('Dominant_Topic')
for i, grp in sent_topics_outdf_grpd:
sent_topics_sorteddf_mallet = pd.concat([sent_topics_sorteddf_mallet,
grp.sort_values(['Perc_Contribution'], ascending=False).head(1)],
axis=0)
# Reset Index
sent_topics_sorteddf_mallet.reset_index(drop=True, inplace=True)
# Format
sent_topics_sorteddf_mallet.columns = ['Topic_Num', "Topic_Perc_Contrib", "Keywords", "Representative Text"]
# Show
sent_topics_sorteddf_mallet.head(10)
# +
from matplotlib import pyplot as plt
from wordcloud import WordCloud, STOPWORDS
import matplotlib.colors as mcolors
cols = [color for name, color in mcolors.TABLEAU_COLORS.items()] # more colors: 'mcolors.XKCD_COLORS'
cloud = WordCloud(stopwords=stop_words,
background_color='white',
width=2500,
height=1800,
max_words=10,
colormap='tab10',
color_func=lambda *args, **kwargs: cols[i],
prefer_horizontal=1.0)
topics = lda_model.show_topics(formatted=False)
fig, axes = plt.subplots(2, 2, figsize=(10,10), sharex=True, sharey=True)
for i, ax in enumerate(axes.flatten()):
fig.add_subplot(ax)
topic_words = dict(topics[i][1])
cloud.generate_from_frequencies(topic_words, max_font_size=300)
plt.gca().imshow(cloud)
plt.gca().set_title('Topic ' + str(i), fontdict=dict(size=16))
plt.gca().axis('off')
plt.subplots_adjust(wspace=0, hspace=0)
plt.axis('off')
plt.margins(x=0, y=0)
plt.tight_layout()
plt.show()
# +
from collections import Counter
topics = lda_model.show_topics(formatted=False)
data_flat = [w for w_list in text for w in w_list]
counter = Counter(data_flat)
out = []
for i, topic in topics:
for word, weight in topic:
out.append([word, i , weight, counter[word]])
df = pd.DataFrame(out, columns=['word', 'topic_id', 'importance', 'word_count'])
# Plot Word Count and Weights of Topic Keywords
fig, axes = plt.subplots(2, 2, figsize=(16,10), sharey=True, dpi=160)
cols = [color for name, color in mcolors.TABLEAU_COLORS.items()]
for i, ax in enumerate(axes.flatten()):
ax.bar(x='word', height="word_count", data=df.loc[df.topic_id==i, :], color=cols[i], width=0.5, alpha=0.3, label='Word Count')
ax_twin = ax.twinx()
ax_twin.bar(x='word', height="importance", data=df.loc[df.topic_id==i, :], color=cols[i], width=0.2, label='Weights')
ax.set_ylabel('Word Count', color=cols[i])
ax_twin.set_ylim(0, 0.030); ax.set_ylim(0, 3500)
ax.set_title('Topic: ' + str(i), color=cols[i], fontsize=16)
ax.tick_params(axis='y', left=False)
ax.set_xticklabels(df.loc[df.topic_id==i, 'word'], rotation=30, horizontalalignment= 'right')
ax.legend(loc='upper left'); ax_twin.legend(loc='upper right')
fig.tight_layout(w_pad=2)
fig.suptitle('Word Count and Importance of Topic Keywords', fontsize=22, y=1.05)
plt.show()
# +
from matplotlib.patches import Rectangle
def sentences_chart(lda_model=lda_model, corpus=mycorpus, start = 0, end = 13):
corp = corpus[start:end]
mycolors = [color for name, color in mcolors.TABLEAU_COLORS.items()]
fig, axes = plt.subplots(end-start, 1, figsize=(20, (end-start)*0.95), dpi=160)
axes[0].axis('off')
for i, ax in enumerate(axes):
if i > 0:
try:
corp_cur = corp[i-1]
topic_percs, wordid_topics, wordid_phivalues = lda_model[corp_cur]
word_dominanttopic = [(lda_model.id2word[wd], topic[0]) for wd, topic in wordid_topics]
ax.text(0.01, 0.5, "Doc " + str(i-1) + ": ", verticalalignment='center',
fontsize=16, color='black', transform=ax.transAxes, fontweight=700)
# Draw Rectange
topic_percs_sorted = sorted(topic_percs, key=lambda x: (x[1]), reverse=True)
ax.add_patch(Rectangle((0.0, 0.05), 0.99, 0.90, fill=None, alpha=1,
color=mycolors[topic_percs_sorted[0][0]], linewidth=2))
word_pos = 0.06
for j, (word, topics) in enumerate(word_dominanttopic):
if j < 14:
ax.text(word_pos, 0.5, word,
horizontalalignment='left',
verticalalignment='center',
fontsize=16, color=mycolors[topics],
transform=ax.transAxes, fontweight=700)
word_pos += .009 * len(word) # to move the word for the next iter
ax.axis('off')
ax.text(word_pos, 0.5, '. . .',
horizontalalignment='left',
verticalalignment='center',
fontsize=16, color='black',
transform=ax.transAxes)
except:pass
plt.subplots_adjust(wspace=0, hspace=0)
plt.suptitle('Sentence Topic Coloring for Documents: ' + str(start) + ' to ' + str(end-2), fontsize=22, y=0.95, fontweight=700)
plt.tight_layout()
plt.show()
sentences_chart()
# +
from sklearn.manifold import TSNE
from bokeh.plotting import figure, output_file, show
from bokeh.models import Label
from bokeh.io import output_notebook
import numpy as np
# Get topic weights
topic_weights = []
for i, row_list in enumerate(lda_model[mycorpus]):
topic_weights.append([w for i, w in row_list[0]])
# Array of topic weights
arr = pd.DataFrame(topic_weights).fillna(0).values
# Keep the well separated points (optional)
arr = arr[np.amax(arr, axis=1) > 0.35]
# Dominant topic number in each doc
topic_num = np.argmax(arr, axis=1)
# tSNE Dimension Reduction
tsne_model = TSNE(n_components=2, verbose=1, random_state=0, angle=.99, init='pca')
tsne_lda = tsne_model.fit_transform(arr)
# Plot the Topic Clusters using Bokeh
output_notebook()
n_topics = 4
mycolors = np.array([color for name, color in mcolors.TABLEAU_COLORS.items()])
plot = figure(title="t-SNE Clustering of {} LDA Topics".format(n_topics),
plot_width=900, plot_height=700)
plot.scatter(x=tsne_lda[:,0], y=tsne_lda[:,1], color=mycolors[topic_num])
show(plot)
# -
import nltk
import urllib
from gensim.models import Word2Vec
import bs4 as bs
import re
source = urllib.request.urlopen("https://en.wikipedia.org/wiki/Global_warming")
soup = bs.BeautifulSoup(source, "lxml")
text = ""
for x in soup.find_all("p"):
text+=x.text
text = nltk.sent_tokenize(text)
len(text)
def my_tokenizer(s):
s = s.lower()
tokens = nltk.tokenize.word_tokenize(s)
tokens = [t for t in tokens if len(t)>2] #remove words lesser than 2 in length
tokens = [wordnet_lemmatizer.lemmatize(t) for t in tokens] #lemmatize words
tokens = [t for t in tokens if t not in custom] #remove stopwords and punctuation
tokens = [t for t in tokens if not any(c.isdigit() for c in t)] # remove digits
return tokens
text = [my_tokenizer(s) for s in text]
# +
# text = [" ".join(x) for x in text]
# -
len(text)
text[:4]
model = Word2Vec(text, min_count = 1)
words = model.wv.vocab
words
vector = model.wv["global"]
vector.shape
similar = model.wv.most_similar("global")
similar
| 14,973 |
/downloaded_kernels/loan_data/kernel_174.ipynb | 40c5abf8ee1838fc2b6160b1c59ab21ba3bbc356 | [
"MIT"
] | permissive | jupste/wranglesearch | https://github.com/jupste/wranglesearch | 0 | 0 | null | null | null | null | Jupyter Notebook | false | false | .py | 13,944 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# +
#Natural Language Processing
# -
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
dataset = pd.read_csv('Restaurant_Reviews.tsv', delimiter = '\t', quoting = 3)
#quoting = 3, for ignoring quotes in dataset
dataset
#Cleaning the texts
import re
review = re.sub('[^a-zA-Z]',' ', dataset['Review'][0])
review
review = review.lower()
review
#removing irrelevant words
import nltk
nltk.download('stopwords')
review = review.split()
review
from nltk.corpus import stopwords
review = [word for word in review if not word in set(stopwords.words('english'))]
#set fxn sed to increase speed, as scanning a set is faster than scanning a list
review
#Stemming ~ taking the root of the word
from nltk.stem.porter import PorterStemmer
ps = PorterStemmer()
review = [ps.stem(word) for word in review if not word in set(stopwords.words('english'))]
review
#joining back the list words into string separated by space
review = ' '.join(review)
review
corpus = []
#corpus means collection of texts pf same type
for i in range (0, 1000):
review = re.sub('[^a-zA-Z]', ' ', dataset['Review'][i])
review = review.lower()
review = review.split()
ps = PorterStemmer()
review = [ps.stem(word) for word in review if not word in set(stopwords.words('english'))]
review = ' '.join(review)
corpus.append(review)
corpus
#Creating the bag of words model
#Creating columns for each unique word in corpus
from sklearn.feature_extraction.text import CountVectorizer
cv = CountVectorizer()
X = cv.fit_transform(corpus).toarray()
X
np.shape(X)
#too much sparsity is there, therefore we will modify our model to remove it
cv1 = CountVectorizer(max_features = 1500)#To find out first 1500 most frequently occur words
X = cv1.fit_transform(corpus).toarray()
#Creating dependent variable vector
y = dataset.iloc[:, 1].values
y
#training the model
#Generally, we use Naive Bayes, Decsion Tree and Random forest classification for Natural Language Processing
#Naive Bayes Classification
from sklearn.cross_validation import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size = 0.20, random_state = 0)
from sklearn.naive_bayes import GaussianNB
classifier = GaussianNB()
classifier.fit(X_train, y_train)
y_pred = classifier.predict(X_test)
y_pred
from sklearn.metrics import confusion_matrix
cm = confusion_matrix(y_test, y_pred)
cm
# +
#54 incorrect predictions out of 200 reviews
# -
accuracy = (55+91)/200
accuracy
| 2,761 |
/hw9/mahoto_sasakihw9.ipynb | 0b9e5abb0d839dbaa4e0296e22c8fc526081eee4 | [] | no_license | MoMoney14/CSE217A | https://github.com/MoMoney14/CSE217A | 0 | 4 | null | null | null | null | Jupyter Notebook | false | false | .py | 265,138 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Homework 9: Feature Engineering
#
# Name: Mahoto Sasaki
#
# Student ID: 467695
#
# Collaborators:
#
# ## Instructions
#
# In this homework, we will be exploring a more applications of feature engineering (both generation adn selection) for supervised machine learning. It will be helpful to review **Lab 9 (Feature Generation)** first. Some of the things we ask you to do in this homework are explained in the lab. In general, you should feel free to import any package that we have previously used in class. Ensure that all plots have the necessary components that a plot should have (e.g. axes labels, a title, a legend).
#
# Furthermore, in addition to recording your collaborators on this homework, please also remember to cite/indicate all external sources used when finishing this assignment. This includes peers, TAs, and links to online sources. Note that these citations will not free you from your obligation to submit your _own_ code and write-ups, however, they will be taken into account during the grading and regrading process.
#
# ### Submission instructions
# * Submit this python notebook including your answers in the code cells as homework submission.
# * **Feel free to add as many cells as you need to** — just make sure you don't change what we gave you.
# * **Does it spark joy?** Note that you will be partially graded on the presentation (_cleanliness, clarity, comments_) of your notebook so make sure you [Marie Kondo](https://lifehacker.com/marie-kondo-is-not-a-verb-1833373654) your notebook before submitting it.
# ## 1. Building a Classifier with Generated Features
#
# In the lab, we used a neural network as a feature extractor. We saw that we could use the $k$-means algorithm to cluster images of cats and dogs with their extracted features. We also saw that we could use $k$-nearest neighbors to build a retrieval system, which was capable of taking a new image of a cat and dog, extract its features, and then look up the most similar images in the retrieval system.
#
# Today, we will try to take this a step farther. Instead of just returning the most similar images, we will try to classify a new image. To do this we will try to use Logistic Regression.
# ### Checking the Data Location
# +
from os import listdir
base_dir = '../Lab9/utility/data'
assert 'PetImages' in listdir(base_dir), f'Couldn\'t find `PetImages` in the `{base_dir}` directory of this lab'
# -
# ### Building Up a Dataset
#
# As you know from the lab, we have 25,000 images in our dataset. There are _way_ too many images for a homework based on feature extraction to be feasible if we used all of them. Instead, we will get the paths of all of the images and sample 400 of them.
# +
classes = ['Dog', 'Cat']
dog_paths = [f'{base_dir}/PetImages/Dog/{image}' for image in listdir(f'{base_dir}/PetImages/Dog')]
cat_paths = [f'{base_dir}/PetImages/Cat/{image}' for image in listdir(f'{base_dir}/PetImages/Cat')]
# -
dog_paths[:5]
# ### Problem 1.1
#
# Next, we will want to sample from these image paths to build our subset. However, we will want to make sure that our dataset is balanced with equal representation of each class.
# **Write-up!** Why is this important?
# + active=""
# # your response here
# Our dataset should be balanced with equal representation for each class becasue we want the same sample size for each class to make comparisons.
# -
# **Try this!** In the cell below, use the provided `choice` function, which is the same as [`np.random.choice`](https://www.numpy.org/devdocs/reference/generated/numpy.random.choice.html?highlight=choice#numpy.random.choice) with an added `random_state` keyword argument, on both `dog_paths` and `cat_paths` to produce a list of `paths` that contains the paths of images we will use in our dataset. Additionally, please prepare the `labels` for these images. Set the `random_state` to 5.
# +
import numpy as np
def choice(*args, random_state=None, **kwargs):
'''
Has the same function interface as np.random.choice
except with the addition of a random_state keyword argument (kwarg)
'''
return np.random.RandomState(seed=random_state).choice(*args, **kwargs)
# +
# your code here
labels = np.concatenate((np.zeros(200), np.ones(200)))
#https://stackoverflow.com/questions/9775297/append-a-numpy-array-to-a-numpy-array
dog_paths = choice(dog_paths, 200, random_state=5)
cat_paths = choice(cat_paths,200, random_state=5)
paths = np.concatenate((dog_paths, cat_paths))
paths = paths.tolist()
print(type(paths))
assert len(paths) == 400, 'Expected 400 image paths in PATHS'
assert labels.shape == (400,), 'Expected 400 labels'
assert labels.sum() == 200, 'Expected 200 [0, 1] labels for each class'
# -
# ### Extracting Features
#
# Now that we have the paths of the images we want to use, the next step in our dataset preparation is to extract the features of these images.
# ### Problem 1.2
#
# To accomodate for different degrees of completion on the lab, we have provided an `preprocess_image` and `extract_features` functions for you to use.
#
# **Try This!** Investigate the `preprocess_image` and `extract_features` functions in `utility/util.py` and use them to produce feature vectors for each image. `Hint` You will need to provide the `preprocess_fn` and the `extraction_model` to `extract_features`.
# +
from utility.util import extract_features
# your code here
from utility.util import preprocess_image
from tensorflow.keras.models import Model
from tensorflow.keras.applications.vgg16 import preprocess_input
from tensorflow.keras.applications.vgg16 import VGG16
base_model = VGG16(weights='imagenet')
layer_name = 'flatten'
extracted_features = extract_features(base_model, preprocess_input, paths)
# -
# ### Creating Our Model!
#
# Now we are ready to start the machine learning part of this section. The first thing we will need to do is to create a train/test split of our data set.
# ### Problem 1.3
#
# **Try this!** In the cell below, create a train/test split of our data set with a test set proportion of `0.25` and a `random_state` of `8`. Additionally, stratify your split with the labels you produced earlier. Finally, also refer to [this StackOverflow answer](https://stackoverflow.com/a/35622967) to get the indices of each point in the training and test sets in the original data; store these as `idx_train` and `idx_test`.
# your code here
from sklearn.model_selection import train_test_split
indices = range(len(extracted_features))
X_train, X_test, y_train, y_test, idx_train, idx_test = train_test_split(extracted_features, labels, indices, test_size=0.25, random_state=8, stratify=labels)
# ### Problem 1.4
#
# Let's build a Logistic Regression classifier with our extracted featureset.
#
# **Try this!** In the cell below, create and fit a new `LogisticRegression` model with `random_state=11` and the training set. Then, evaluate the model on _both_ the training and testing sets. If you get a warning, adjust your implementation accordingly to resolve it.
# +
# your code here
from sklearn.linear_model import LogisticRegression
logisticModel = LogisticRegression(random_state=11, solver='liblinear')
logisticModel.fit(X_train, y_train)
y_pred = logisticModel.predict(X_test)
y_model = logisticModel.predict(X_train)
#train test accuracy
from sklearn.metrics import accuracy_score
testingAccuracy = accuracy_score(y_test, y_pred)
print(testingAccuracy)
#how to get testing accuracy?
trainingAccuracy = accuracy_score(y_train, y_model)
print(trainingAccuracy)
testing_acc = testingAccuracy, trainingAccuracy
# -
testing_acc
# **Write-up** What were the training and testing accuracies for this model? What do each of these values mean? What do they tell us about our model?
# + active=""
# # your response here
# The training accuracy was 0.98 and this value indicates the correct percentage of classifications for the model on the training data. The testing accuracy was 0.92 and this value indicates the correct percentage of classifications for the model on the testing data. This shows that our model should be pretty good at predicting the outcomes of new data.
# -
# ### Classifying Cats and Dogs!
#
# Now that we have trained and evaluated our model, let's get to the fun part: looking at pictures of cats and dogs!
# ### Problem 1.5
#
# **Try this!** Use the `choice` function provided in [Problem 1.1](#Problem-1.1) with a `random_state` of 14 to sample the feature vectors of five images from our testing set. Then, use the Logistic regression model we trained in [Problem 1.4](#Problem-1.4) to classify each of the images, reporting both the class label, $\hat y$, and the probability of that class, $P(y = \hat y \mid x)$. Record the indices of these images in `image_indices`.
# +
# your code here
# print(len(idx_train))
# print(len(idx_test))
# print(len(X_test))
# print(len(X_train))
# print(len(y_train))
# print(len(y_test))
print(idx_test)
print(len(X_test))
print(len(idx_test))
print()
images = choice(idx_test, 5,random_state=14)
image_indices = []
for j in images:
for idx, i in enumerate(idx_test):
if i==j:
image_indices.append(idx)
test_images =[]
for i in image_indices:
test_images.append(X_test[i])
for i in image_indices:
classLabel = logisticModel.predict(test_images)
print(classLabel)
probability = logisticModel.predict_proba(test_images)
print(probability)
# -
# Let's check how our model did.
#
# **Try this!** Use the `show_image` function to display each image in `image_indices`.
# +
from utility.util import show_image
for index in image_indices:
path = paths[idx_test[index]]
show_image(path)
# -
# ### Problem 1.Not a Problem
#
# 
#
# Okay. Moving on.
# ## 2. Automatic Feature Selection
# In lab 9 and in the previous section, we have seen how we can use a pretrained neural network model to extract features. However, it is often not possible or not feasible to use a neural network to do this. Additionally, we have also seen the problems with having too many features and too many model paramenters.
#
# In this section, we will cover two different methods of selecting features, or in other words, removing some features, to hopefully improve the accuracy of the resulting model by accounting for the effects of overfitting.
# ### Loading the Data
#
# First, we will load the breast cancer dataset and its 30 features.
# +
from sklearn.datasets import load_breast_cancer
cancer = load_breast_cancer()
X, y = cancer.data, cancer.target
# -
# ### Adding Some Noise
#
# We want to simulate what happens when we have too many features, many of which are redundant or are just noise. Let's add 30 features that are random noise.
noise = np.random.RandomState(28).normal(size=X.shape)
X_noise = np.hstack([X, noise])
# ### Creating Training, Validation, and Testing Sets
#
# We then split the data into test and train sets, as well as an additional validation set to compare the final results of the models generated.
# ### Problem 2.1
#
# In the following cell, produce a test set with 20% of the `X_noise`, then use 10% of the remaining data as a validation set. The data left will be our training set. Stratify by the target variable (remember how this data set was unbalanced). Also, use a `random_state` of 13.
from sklearn.model_selection import train_test_split
# +
# your code here
X_train, X_test, y_train, y_test = train_test_split(X_noise, y, test_size=0.2, random_state=13, stratify=y)
X_train, X_val, y_train, y_val = train_test_split(X_train, y_train, test_size=0.10, random_state=13, stratify=y_train)
# -
# ### Model-Based Feature Selection
# In model-based feature selection, a model (that could be different from the one used to train the data) selects the features that are most important by looking at the `feature_importances_` attribute that decision tree models provide. It accounts for interactions between features as well, unlike some other methods that only look at features individually.
# +
from sklearn.feature_selection import SelectFromModel
from sklearn.ensemble import RandomForestClassifier
model = RandomForestClassifier(n_estimators=100, random_state=28)
select = SelectFromModel(model, threshold="median")
# -
# We use a random forest classifier with 100 trees to compute the feature importances, and select half of the 60 features (this is the 'threshold="median"' parameter. A random forest classifier uses multiple decision trees on different subsets of the data to reduce the tendency for trees to overfit.
select.fit(X_train, y_train)
X_train_selected = select.transform(X_train)
print(f' Noisy Features: {X_train.shape}')
print(f'Selected Features: {X_train_selected.shape}')
# To see what has been selected, we can plot an array of the indices of the features. A black index indicates a selected feature.
# +
import matplotlib.pyplot as plt
from utility.util import configure_plots
# run this twice!
configure_plots()
mask = select.get_support()
plt.matshow(mask.reshape(1, -1), cmap='gray_r')
plt.xlabel("Sample index")
plt.yticks(())
plt.show()
# -
# ### Problem 2.2
#
# Let's test the original set of features and the selected ones to see the difference, using a logistic regression.
#
# **Try This!** Create two new `LogisticRegression` models. Fit the first with the noisy training data `X_train` and the feature-selected training set `X_train_selected`. Then, use the selector to transform the noisy test set `X_test` and store the result in `X_test_selected`. Finally, evaluate both models with their respective test sets and report the results. Use a `random_state` of 15 and set the solver for both models to be `liblinear`.
# +
from sklearn.linear_model import LogisticRegression
# your code here
model1 = LogisticRegression(random_state = 15, solver='liblinear').fit(X_train, y_train)
model2 = LogisticRegression(random_state = 15, solver='liblinear').fit(X_train_selected, y_train)
X_test_selected = select.transform(X_test)
#calculate accuracies
y_pred1 = model1.predict(X_test)
testingAccuracy1 = accuracy_score(y_test, y_pred1)
print(testingAccuracy1)
y_pred2 = model2.predict(X_test_selected)
testingAccuracy2 = accuracy_score(y_test, y_pred2)
print(testingAccuracy2)
# -
# ### Iterative Feature Selection
# In iterative feature selection, a series of models are built, removing or building up one feature at a time, as opposed to constructing one model from which the entire selection is made. A particular kind of this is this is [**recursive feature elimination** (RFE)](https://scikit-learn.org/stable/modules/generated/sklearn.feature_selection.RFE.html), which starts with all features, builds a model, and discards the least important feature according to the model. Then a new model is built using all but the discarded feature, and so on until only a prespecified number of features are left. For this to work, the model used for selection needs to provide some way to determine feature importance, as was the case for the model-based selection. Here, we use the same random forest model that we used earlier.
#
# In this code, we will run 40 iterations of RFE, selecting 59 of the 60 features, then 58, and so on, until only 20 are selected. At this point, we expect the accuracy to be somewhat lower, given that we would have to get rid of some of the original 30 features. We also save the features at the iteration that selects 30, to compare to the ones selected above.
# +
from sklearn.feature_selection import RFE
from IPython import display
scores, masks = [], []
#set up axes for plot
fig, (ax1, ax2) = plt.subplots(2, 1)
for number_to_remove in range(1, 50):
model = RandomForestClassifier(n_estimators=100, random_state=28)
selector = RFE(model, n_features_to_select=60-number_to_remove)
selector.fit(X_train, y_train)
# create and save masks
masks.append(selector.get_support())
X_train_rfe = selector.transform(X_train)
X_test_rfe = selector.transform(X_test)
model = LogisticRegression(solver='liblinear')
model.fit(X_train_rfe, y_train)
score = model.score(X_test_rfe, y_test)
scores.append(score)
# update plot
ax1.clear()
ax1.plot(scores, 'b-')
ax1.set_xlim(-1, 51)
ax1.set_ylim(0.90, 0.95)
ax1.set_xlabel('Number of Features to Remove')
ax1.set_ylabel('Accuracy')
ax2.matshow(selector.get_support().reshape(1, -1), cmap='gray_r')
ax2.set_yticks(())
ax2.set_xlabel('Feature Index')
display.clear_output(wait=True)
display.display(fig)
# -
# As you can see, feature selection has a generally positive impact on this data set, given that we added 30 extra features of noise. However, it is a tool you should keep in mind when combatting overfitting. But, beware of the impact of removing too many features too.
# ### Problem 2.3
#
# **Write-up!** Given the plots produced above, how many features should we remove using the RFE algorithm? Why? Additionally, why does each successive iteration take longer than the last and what drawbacks this might have in real world applications?
# + active=""
# # your response here
# We should remove 10 or 11 features based on the RFE algorithm because that is the number of features that need to be removed to produce the highest accuracy. Each successive iteration takes longer than the last because if it wants to remove n features it restarts by starting from the beginning of the data to n features. Therefore, as the number of features to remove increases, the time it takes to process the data get longer. The drawback of this algorithm is that there are performance issues. In the real world, you do not want the algorithm to take this long because it decreases efficiency.
#
# -
# **Try this!** In the cell below, store the mask from `masks` from the iteration number that you chose above in `best_mask`. We will plot the features selected during this iteration.
# +
# your code here
best_mask = masks[10]
plt.matshow(best_mask.reshape(1, -1), cmap='gray_r')
plt.xlabel("Sample index")
plt.yticks(())
# -
# ### Problem 2.4
#
# **Write-up!** Consider both of the methods we just saw: model-based feature selection and iterative feature selection. How did the resulting, feature-selected, datasets perform relative to the noisy ones? What are the differences between the two methods? What are the pros and cons of each method? Is there one that you should prefer over the other?
# + active=""
# # your response here
# The datasets performed better than the noisy ones.
#
# The differences between the two methods are that one is an iterative approach while the other uses a decision tree.
#
# The pros for the model-based feature datasets are that you can filter out the unnecessary features or the features that do not contribute a lot to the target variable. The cons for the feature-selected datasets are that not all models can compute feature importance.
#
# The pros for the iterative feature datasets are that you can filter out the unnecessary features or the features that do not contribute a lot to the target variable. The con is that an iterative based approach takes a lot of time to compute and requires validation sets so you will lose some training data.
#
# Generally, it depends on the situation of when to use the feature-selected dataset over the iterative feature selection because they might want to optimize for efficiency.
| 19,792 |
/Naver_Headline.ipynb | ef38a8edf538787efe2f0c42ff3d96f26ffa745d | [] | no_license | minjae2271/web_crawl | https://github.com/minjae2271/web_crawl | 0 | 0 | null | null | null | null | Jupyter Notebook | false | false | .py | 15,013 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# +
from bs4 import BeautifulSoup as bs
with open('/Users/minjae/Desktop/2.html') as fp:
soup = bs(fp, 'html.parser')
print(soup)
#print(soup.prettify)
# +
#tag parse 하기
# -
soup.h1
soup.h1.name
soup.h1.string
soup.div
soup.img
soup.img.parent
soup.img.parent.name
soup.p
soup.a
#태그검색
find() : 조건에 맞는 태그를 하나 가져옴
find_all() : 조건에 맞는 태그를 모두 가져옴
# +
soup_find = soup.find('div')
print(soup_find)
# 위의 파싱과 동일한 결과
# +
soup_find_all = soup.find_all('div')
print(soup_find_all)
#리스트 형태로 반환
# -
find_by_class = soup.find_all('div',{'class':'logos'})
print(find_by_class)
find_by_id = soup.find_all('a',{'id':'python'})
print(find_by_id)
soup.find('a').get('href')
# +
#soup.find('a').string
soup.find('a').get_text()
# +
site_url = soup.find_all('a')
#print(site_url)
for url in site_url:
print(url.get('href'))
#print(url.name)
# -
# #urllib : 웹사이트에서 쉽게 정보를 가져올 수 있게 해주는 라이브러리
# +
from urllib import request
from urllib import parse
from bs4 import BeautifulSoup as bs
res = request.urlopen("https://www.naver.com/")
# -
print(res)
# +
res.code
#200 : ㅇㅋ
#404 : ㄴㄴ
# -
res.peek()
html = res.read()
bs(html,'html.parser')
# 이수안 컴퓨터 연구소 : 네이버 뮤직 100
# +
from urllib import request
from urllib import parse
from bs4 import BeautifulSoup as bs
url = 'https://music.naver.com/listen/top100.nhn?domain=DOMESTIC_V2&page=1'
html = request.urlopen(url).read()
soup = bs(html, 'html.parser')
# print(soup)
#<a href="#42644553" class="_title title NPI=a:track,r:2,i:42644553" title="Lovesick Girls"><span class="ellipsis">Lovesick Girls</span></a>
titles = soup.find_all('a',{'class':'_title'})
for title in titles:
print(title.get_text())
# -
# #parser : UTF-8로 변환
search = request.urlopen("https://search.naver.com/search.naver?sm=top_hty&fbm=1&ie=utf8&query=%ED%94%BC%EC%B9%B4%EC%B8%84")
search.code
search = request.urlopen("https://search.naver.com/search.naver?sm=top_hty&fbm=1&ie=utf8&query=꼬부기")
search.code
# +
baseUrl = "https://search.naver.com/search.naver?sm=top_hty&fbm=1&ie=utf8&query="
plusUrl = "꼬부기"
url = baseUrl + plusUrl
print(url)
# +
plusUrl = parse.quote_plus(plusUrl)
url = baseUrl + plusUrl
print(url)
# -
# #프로그래머 김플 스튜디오 강좌
import urllib.request
import urllib.parse
from bs4 import BeautifulSoup as bs
from bs4 import NavigableString, Tag
# +
baseUrl = 'https://search.shopping.naver.com/search/all?where=all&frm=NVSCTAB&query='
plusUrl = input('검색어를 입력하세요:')
url = baseUrl + urllib.parse.quote_plus(plusUrl)
html = urllib.request.urlopen(url).read()
soup = bs(html, 'html.parser')
#__next > div > div.container > div.style_inner__18zZX > div.style_content_wrap__1PzEo > div.style_content__2T20F > ul > div > div:nth-child(1) > li > div > div.basicList_info_area__17Xyo > div.basicList_title__3P9Q7
title = soup.find_all(class_='basicList_link__1MaTN')
print(len(title))
#print(title)
for ele in title:
print(ele.get('title'))
print(ele.get('href'))
print()
# for ele in title:
# if isinstance(ele, NavigableString):
# continue
# if isinstance(ele, Tag):
# print(ele.attrs['title'])
# print(ele.attrs['href'])
# print()
# -
# #Naver 이미지 크롤링
# +
#https://search.naver.com/search.naver?where=image&sm=tab_jum&query=%EC%82%AC%EA%B3%BC
baseUrl = 'https://search.naver.com/search.naver?where=image&sm=tab_jum&query='
plusUrl = input('검색어를 입력하세요')
url = baseUrl + urllib.parse.quote_plus(plusUrl)
html = urllib.request.urlopen(url).read()
soup = bs(html, 'html.parser')
img = soup.find_all(class_='_img')
n = 1
for img_ele in img:
imgUrl = img_ele.get('data-source')
print(imgUrl)
with urllib.request.urlopen(imgUrl) as f:
#바이너리기 때문에 wb
with open('./naver_img/' + plusUrl + str(n) + '.jpg', 'wb') as h:
img = f.read()
h.write(img)
n+=1
print('다운로드 완료!')
# -
# #인스타그램 이미지 크롤링
#
# 사전준비 : chromedriver (https://chromedriver.chromium.org/downloads 상위 버전 다운로드) -> 파이썬파일과 동일 경로에 다운받은 파일 이동
#
# 구글검색창에 instagram 파이썬으로 검색하면 로그인 하지않고 게시물 검색 가능
from urllib.request import urlopen
from urllib.parse import quote_plus
from bs4 import BeautifulSoup
from selenium import webdriver
from selenium.webdriver.common.keys import Keys
import time
# +
#https://www.instagram.com/explore/tags/%EC%9D%84%EC%A7%80%EB%A1%9C%EB%A7%9B%EC%A7%91/
baseUrl = 'https://www.instagram.com/explore/tags/'
plusUrl = input('인스타 검색어를 입력하세요.')
url = baseUrl + quote_plus(plusUrl)
#인스타그램의 웹구조 -> JS기 때문에 selenium 사용
#.Chrome() -> 에러발생하여 절대경로 추가 (driver파일을 usr/bin으로 옮겨도 해결된다고 함)
driver = webdriver.Chrome('/Users/minjae/web_crawl/chromedriver')
driver.get(url)
body = driver.find_element_by_css_selector('body')
for i in range(5):
body.send_keys(Keys.PAGE_DOWN)
time.sleep(.5)
time.sleep(3)
html = driver.page_source
soup = BeautifulSoup(html,'html.parser')
# v1Nh3 kIKUG _bz0w class가 3개 -> .으로 구분
insta = soup.select('.v1Nh3.kIKUG._bz0w')
#출력 확인하는 습관!
print(insta[0])
# n = 1
# for i in insta:
# print("https://www.instagram.com/" + i.a['href'])
# imgUrl = i.select_one('.KL4Bh').img['src']
# with urlopen(imgUrl) as f:
# with open('./insta_img/' + plusUrl + str(n) + 'jpg', 'wb') as h:
# img = f.read()
# h.write(img)
# n+=1
# driver.close()
# -
# #네이버 블로그 크롤링 (여러페이지) -> page 처리가 관건!
# #1번 소스 코드를 수정하여 만들어보자
# #1번 import cell 실행 해주세요
#
# <a href="?date_from=&date_option=0&date_to=&dup_remove=1&nso=&post_blogurl=&post_blogurl_without=&query=%ED%8C%8C%EC%9D%B4%EC%8D%AC&sm=tab_pge&srchby=all&st=sim&where=post&start=51" onclick="return goOtherCR(this,'a=blg.paging&i=&r=6&u='+urlencode(urlexpand(this.href)));">6</a>
#
# <a href="?date_from=&date_option=0&date_to=&dup_remove=1&nso=&post_blogurl=&post_blogurl_without=&query=%ED%8C%8C%EC%9D%B4%EC%8D%AC&sm=tab_pge&srchby=all&st=sim&where=post&start=61" onclick="return goOtherCR(this,'a=blg.paging&i=&r=7&u='+urlencode(urlexpand(this.href)));">7</a>
#
# <a href="?date_from=&date_option=0&date_to=&dup_remove=1&nso=&post_blogurl=&post_blogurl_without=&query=%ED%8C%8C%EC%9D%B4%EC%8D%AC&sm=tab_pge&srchby=all&st=sim&where=post&start=71" onclick="return goOtherCR(this,'a=blg.paging&i=&r=8&u='+urlencode(urlexpand(this.href)));">8</a>
#
# -> 6페이지의 start=51 / 7페이지의 start=61 / 8페이지의 start=71
#
#
# <a class="sh_blog_title _sp_each_url _sp_each_title" href="https://blog.naver.com/0813dcba?Redirect=Log&logNo=221950381025" target="_blank" onclick="return goOtherCR(this, 'a=blg*i.tit&r=1&i=90000003_0000000000000033AD460BE1&u='+urlencode(this.href))" title="푸드엔샵 에콜로 착즙주스 사과즙 대신 챙겨요">푸드엔샵 에콜로 착즙주스 <strong class="hl">사과</strong>즙 대신 챙겨요</a>
# +
plusUrl = urllib.parse.quote_plus(input('검색어를 입력하세요:'))
pageNum = 1
i = input("몇페이지까지 크롤링을 할까요?")
lastPage = int(i) * 10 -9
count = 1
while pageNum<lastPage+1:
url = f'https://search.naver.com/search.naver?date_from=&date_option=10&date_to=&dup_remove=1&nso=&post_blogurl=&post_blogurl_without=&query={plusUrl}&sm=tab_pge&srchby=all&st=sim&where=post&start={pageNum}'
html = urllib.request.urlopen(url).read()
soup = bs(html, 'html.parser')
title = soup.find_all(class_='sh_blog_title')
print(f'--------------{count}페이지 결과입니다')
for ele in title:
print(ele.attrs['title'])
print(ele.attrs['href'])
print()
count+=1
pageNum += 10
| 7,814 |
/stock prediction 2.ipynb | 07860a8b134521c0deb56f3113c83f282bb78cdf | [] | no_license | SameerSinghDudi/Stock-Price-Prediction-using-RNN-LSTM | https://github.com/SameerSinghDudi/Stock-Price-Prediction-using-RNN-LSTM | 0 | 0 | null | null | null | null | Jupyter Notebook | false | false | .py | 5,094,769 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# +
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
from keras.models import Sequential
from keras.layers import LSTM
from keras.layers import Dense
from keras import metrics
from sklearn.preprocessing import MinMaxScaler
from sklearn.model_selection import train_test_split
import yfinance as yf
# -
fb = yf.download('FB', '2015-01-01', '2020-01-01')
fb.head()
fb['close_next_day'] = fb['Close'].shift(-1, axis=0)
fb.head()
df = fb[['High','Low','close_next_day']]
df.head()
high = df['High'].to_numpy()
low = df['Low'].to_numpy()
y = df['close_next_day'].to_numpy()
high = np.delete(high,-1)
low = np.delete(low,-1)
y = np.delete(y,-1)
y.shape
y = y.reshape(1258,1)
y
x = np.vstack((high,low)).T
x
x.shape
y.shape
scaler = MinMaxScaler()
scaler.fit(x)
x = scaler.transform(x)
scaler1 = MinMaxScaler()
scaler1.fit(y)
y = scaler1.transform(y)
x
y
x = x.reshape(1258,1,2)
x
x_train, x_test, y_train, y_test = train_test_split(x,y,test_size = 0.25)
x_train.shape
y_train.shape
# +
model = Sequential()
model.add(LSTM(75, input_shape = (1,2), activation='tanh', recurrent_activation='sigmoid'))
model.add(Dense(1))
model.compile(loss='mse', optimizer = 'adam', metrics = [metrics.mae])
model.summary()
# -
history = model.fit(x_train,y_train, epochs=100)
predict = model.predict(x_test)
# +
plt.figure(1, figsize= (32,16))
plt.plot(predict)
plt.plot(y_test)
plt.legend(['Predicted by Model','Actual Output'])
plt.title('Performance')
plt.show()
# -
original_value = scaler1.inverse_transform(y_test)
len(original_value)
original_value = original_value.reshape(315,)
ov = pd.Series(original_value)
predicted_value = scaler1.inverse_transform(predict)
len(predicted_value)
predicted_value = predicted_value.reshape(315,)
pv = pd.Series(predicted_value)
# +
plt.figure(2, figsize = (160,80))
plt.plot(original_value)
plt.plot(predicted_value)
plt.legend(['Original', 'Predicted'])
plt.show()
# -
result = pd.DataFrame({'Original Value':ov, 'Predicted Value': pv})
df
googl = yf.download('GOOGL', '2015-01-01','2020-01-01')
googl['close_next_day'] = googl['Close'].shift(-1)
high = googl['High'].to_numpy()
low = googl['Low'].to_numpy()
y = googl['close_next_day'].to_numpy()
high = np.delete(high,-1)
low = np.delete(low,-1)
y = np.delete(y,-1)
x = np.vstack((high,low)).T
x.shape
y = y.reshape(1258,1)
scaler = MinMaxScaler()
scaler.fit(x)
x = scaler.transform(x)
scaler1 = MinMaxScaler()
scaler1.fit(y)
y = scaler1.transform(y)
x = x.reshape(1258,1,2)
y.shape
predict = model.predict(x)
# +
plt.figure(3, figsize=(32,16))
plt.plot(y)
plt.plot(predict)
plt.legend(['original','predict'])
plt.title('Model Performance')
plt.show()
# -
result.head()
result['error'] = result['Original Value'] - result['Predicted Value']
result.head()
result.describe()
ocalhost:8080/", "height": 35}
chars = sorted(list(set(text)))
print("Elofordulo karakterek szama:", len(chars))
# + id="jpHjBZ0gHI2k" colab_type="code" outputId="9fb7c72b-9211-433e-8bd8-e2d472db7bab" colab={"base_uri": "https://localhost:8080/", "height": 849}
chars
# + id="NBq1tcwNHQ-S" colab_type="code" colab={}
char_indices = dict((c,i) for i,c in enumerate(chars))
# + id="cF_EIRCMHaMt" colab_type="code" outputId="80d76f9c-5e32-4a8e-8df3-1d12577e2ff0" colab={"base_uri": "https://localhost:8080/", "height": 55}
print(char_indices)
# + id="x-9kC34uHfzZ" colab_type="code" outputId="8d5bf8f6-e951-4956-c74e-57473dd13a6a" colab={"base_uri": "https://localhost:8080/", "height": 55}
indices_char = dict((i,c) for i,c in enumerate(chars))
print(indices_char)
# + [markdown] id="lWX5G_auHsQ7" colab_type="text"
# Szöveg numerikussá alakítása és ablakokra való vágása:
# + id="jcPqqEuHHwcn" colab_type="code" colab={}
maxlen = 40
step = 3
sentences = []
next_chars = []
for i in range(0, len(text)-maxlen, step):
sentences.append(text[i:i+maxlen])
next_chars.append(text[i+maxlen])
# + id="HmN3Gw7aIMlF" colab_type="code" outputId="25b8466f-dbd3-4822-83c4-b90720561a7f" colab={"base_uri": "https://localhost:8080/", "height": 17177}
sentences
# + id="lTFrjNhJIXWW" colab_type="code" outputId="a34f04ec-9f36-4c1a-c235-2eb7700285e5" colab={"base_uri": "https://localhost:8080/", "height": 34}
print("Tanító adatbázis hossza:", len(sentences))
# + id="frokycQtIgBd" colab_type="code" outputId="7bace6e2-b164-4012-fce0-04f854d67cbf" colab={"base_uri": "https://localhost:8080/", "height": 54}
print(next_chars)
# + id="6QlB3VaIIqs7" colab_type="code" colab={}
X = np.zeros((len(sentences), maxlen, len(chars)))
Y = np.zeros((len(sentences), len(chars)))
for i, sentence in enumerate(sentences):
for t, char in enumerate(sentence):
X[i,t,char_indices[char]] = 1
Y[i, char_indices[next_chars[i]]] = 1
# + id="iq-Jgwc-LHPA" colab_type="code" outputId="72674599-5f87-4864-c350-fea6d448d20f" colab={"base_uri": "https://localhost:8080/", "height": 51}
print(X.shape)
print(Y.shape)
# + id="V9Apw8BqLdds" colab_type="code" colab={}
model = Sequential()
model.add(LSTM(32, recurrent_dropout=0.3, input_shape=(X.shape[-2], X.shape[-1],)))
model.add(Dense(len(chars)))
model.add(Activation('softmax'))
model.compile(loss='categorical_crossentropy', optimizer='adam')
# + id="nKCwrmAhSR8s" colab_type="code" outputId="c211a303-db65-4056-dacb-58f96a51607a" colab={"base_uri": "https://localhost:8080/", "height": 240}
model.summary()
# + id="qkwiryrEMrxM" colab_type="code" outputId="29ecf777-8bcd-48c0-ae50-973dae06369f" colab={"base_uri": "https://localhost:8080/", "height": 634}
def sample(preds, temperature=1.0):
preds = np.asarray(preds).astype(np.float64)
preds = np.log(preds) / temperature
exp_preds = np.exp(preds)
preds = exp_preds / np.sum(exp_preds)
probas = np.random.multinomial(1,preds,1)
return np.argmax(probas),preds
fake_preds = [0.1, 0.2, 0.3, 0.15, 0.25]
for temp in [0.1, 0.5, 1, 2, 4]:
print("Temp:",temp)
print(fake_preds)
proba, preds = sample(fake_preds,temp)
print(preds)
print(proba)
print("\n\n")
# + id="2XurhIEQSz_R" colab_type="code" outputId="e993075c-05c3-4e77-d6ff-e1d2d120d429" colab={"base_uri": "https://localhost:8080/", "height": 5111}
start_index=random.randint(0,len(text)-maxlen-1)
for iteration in range(1,50):
print("Iteracio szama:", iteration)
model.fit(X,Y, batch_size=256, epochs=10)
for temp in [0.1,0.5,1.0,1.2,2]:
print("Temp:",temp)
generated_text = ''
sentence = text[start_index:start_index+maxlen]
generated_text = sentence
for i in range(200):
x=np.zeros((1,maxlen,len(chars)))
for t,char in enumerate(sentence):
x[0,t,char_indices[char]]=1
preds = model.predict(x, verbose=0)[0]
next_index, _ = sample(preds, temp)
next_char = indices_char[next_index]
generated_text += next_char
sentence = sentence[1:] + next_char
sys.stdout.write(next_char)
sys.stdout.flush()
print("\n")
# + [markdown] id="r_vpfgyrxZur" colab_type="text"
# # 2018.10.25. - 1D CNN
# + id="3odYx4nFxY86" colab_type="code" colab={}
import numpy as np
import matplotlib.pyplot as plt
# %matplotlib inline
num_samples=1000
lp = np.linspace(-10*np.pi, 10*np.pi, num_samples)
x = 30*(np.random.rand(num_samples)-0.5)+np.sin(lp)*3+np.cos(np.random.rand(num_samples)+lp*2)+np.cos(np.pi/2+lp*4)*15 + np.log(np.abs(lp))
# + id="zOCeHECzx83b" colab_type="code" outputId="1e87f1f2-a7cc-4497-d75d-6f5806b589ed" colab={"base_uri": "https://localhost:8080/", "height": 35}
print(x.shape)
# + id="-KvL5wIhyDrl" colab_type="code" outputId="ba34aa8e-306c-407d-e26f-a0d2b5be8173" colab={"base_uri": "https://localhost:8080/", "height": 610}
plt.figure(figsize=(10,10))
plt.plot(lp,x)
# + id="Dt2F0aF82BIk" colab_type="code" colab={}
window_size=20
valid_split=0.3
test_split =0.2
from keras.models import Sequential
from keras.layers.core import Activation, Dense, Flatten
from keras.optimizers import SGD
from keras.callbacks import ModelCheckpoint, EarlyStopping
from sklearn import preprocessing
from keras.layers.convolutional import Convolution1D, MaxPooling1D
from keras.layers.convolutional import Conv1D
def make_1d_convnet(window_size, filter_length, nb_input_series=1, nb_output=1, nb_filter=4):
model = Sequential()
# Conv1D(filters=nb_filter, kernel_size=filter_length, activation='relu',...)
model.add(Convolution1D(nb_filter=nb_filter, filter_length=filter_length, activation='relu',
input_shape=(window_size, nb_input_series, )))
model.add(MaxPooling1D()) # filter:2, stride:2
model.add(Convolution1D(nb_filter=nb_filter, filter_length=filter_length, activation='relu'))
model.add(MaxPooling1D()) # filter:2, stride:2
model.add(Flatten())
model.add(Dense(1,activation='linear'))
model.compile(loss='mse', optimizer='adam', metrics=['mae'])
return model
#model=make_1d_convnet(window_size=window_size, filter_length=5)
# + id="pjky5ovu9UNS" colab_type="code" colab={}
def make_timeseries_innstance(timeseries, window_size):
timeseries = np.asarray(timeseries)
assert 0 < window_size < timeseries.shape[0], "Out of range"
X = np.atleast_3d(np.array([timeseries[start:start+window_size] for start in range(0,timeseries.shape[0]-window_size)]))
assert len(X.shape) == 3, "Dimension error"
Y = timeseries[window_size:]
return X, Y
# + id="treQSE10_Bnz" colab_type="code" colab={}
def eval_timeseries(timeseries, window_length):
filter_length = 5
nb_filter = 4
timeseries = np.atleast_2d(timeseries)
if timeseries.shape[0]==1:
timeseries = timeseries.T
nb_samples, nb_series = timeseries.shape
model=make_1d_convnet(window_size=window_size,
filter_length=filter_length,
nb_input_series=nb_series,
nb_filter=nb_filter)
print(model.summary())
X, Y = make_timeseries_innstance(timeseries, window_size)
test_size = int(nb_samples*(1-test_split))
valid_size = int(nb_samples*(1-valid_split-test_split))
X_train, Y_train = X[:valid_size], Y[:valid_size]
X_valid, Y_valid = X[valid_size:test_size], Y[valid_size:test_size]
X_test, Y_test = X[test_size:], Y[test_size:]
##$$$$$ NE FELEJTSETEK EL A STANDARADIZALAST $$$$$$
##$$$$$$ NE FELEJTESTEK EL EARLY STOPPINGOT ES MCP-t $$$$
model.fit(X_train, Y_train, epochs=50, validation_data=[X_valid,Y_valid], verbose=2)
##$$$$$ NE FELETSETEK EL AZ MCP-t VISSZATOLTENI $$$$
preds = model.predict(X_test)
plt.figure(figsize=(10,10))
plt.plot(preds, color='r')
plt.plot(Y_test, color='b')
# + id="bU-ROpiqA76X" colab_type="code" outputId="730e3eb4-73ed-4fd5-bac6-c68cd90786ba" colab={"base_uri": "https://localhost:8080/", "height": 2745}
eval_timeseries(x, window_size)
# + id="SPm01W3LB4e0" colab_type="code" outputId="eb807a7c-0464-48ab-c314-c9d2339ccafe" colab={"base_uri": "https://localhost:8080/", "height": 2745}
eval_timeseries(x, window_size)
# + id="Zxy1HPCWDCYV" colab_type="code" outputId="3d477432-06ec-40c8-c0bd-8906b3dceb27" colab={"base_uri": "https://localhost:8080/", "height": 2764}
eval_timeseries(x, window_size)
# + [markdown] id="Ajtvbh3ul58d" colab_type="text"
# # 2018.10.18.
#
# + id="YlZbNuYpmBi4" colab_type="code" outputId="cd199eac-34ee-4a91-da8d-226917be81db" colab={"base_uri": "https://localhost:8080/", "height": 34}
from keras.datasets import mnist
from keras.utils import np_utils
(x_train, y_train), (x_test, y_test) = mnist.load_data()
# + id="Y25GaFu1tJcM" colab_type="code" colab={}
x_train = x_train.reshape(-1,28*28)
x_test = x_test.reshape(-1,28*28)
# + id="NgmFx5nFtQO6" colab_type="code" colab={}
x_train = x_train.astype("float32")
x_test = x_test.astype("float32")
# + id="rXm8EdFBmCG3" colab_type="code" colab={}
x_train = x_train / 255
x_test = x_test / 255
# + id="fXQ2Ms3nmDF8" colab_type="code" colab={}
y_train = y_train.reshape(-1,1)
y_test = y_test.reshape(-1,1)
# + id="TIyfvB3Rtr1p" colab_type="code" colab={}
y_train = np_utils.to_categorical(y_train, 10)
y_test = np_utils.to_categorical(y_test, 10)
batch_size=128
# + id="WTRzKyWpmHtf" colab_type="code" outputId="916750e9-d37c-41ce-e69b-7669fee4d194" colab={"base_uri": "https://localhost:8080/", "height": 54}
'''
import tensorflow as tf
sess = tf.Session()
# bemenet és kimenet definiálása
x = tf.placeholder(tf.float32, shape=[None, x_train.shape[-1]])
y_ = tf.placeholder(tf.float32, shape=[None, y_train.shape[-1]])
# a súlymátrixa és bias vektor definiálása
W = tf.Variable(tf.zeros([x_train.shape[-1],10]))
b = tf.Variable(tf.zeros([y_train.shape[-1]]))
# inicializálás
sess.run(tf.global_variables_initializer())
# a regresszió létrehozása
y = tf.nn.softmax(tf.matmul(x,W) + b)
# a költségfüggvény megadása
cross_entropy = tf.reduce_mean(-tf.reduce_sum(y_ * tf.log(y), reduction_indices=[1]))
# optimációs algoritmus megadása
train_step = tf.train.GradientDescentOptimizer(0.5).minimize(cross_entropy)
# tanítás 1000 epoch, batch_size batch méret
batch_counter=0
for i in range(1000):
train_step.run(session=sess, feed_dict={x: x_train[batch_counter*batch_size:(batch_counter+1)*batch_size], y_: y_train[batch_counter*batch_size:(batch_counter+1)*batch_size]})
batch_counter=batch_counter+1
if (batch_counter*batch_size > len(x_train)):
batch_counter=0
'''
# + id="WwSFtEnTmJKJ" colab_type="code" outputId="e43cc293-e57c-442c-80c6-12e6247941a1" colab={"base_uri": "https://localhost:8080/", "height": 34}
'''
# ez megadja, hogy hányszor találtuk el
# (az argmax megadja a legnagyobb értékhez tartozó indexet a második paraméter dimenziójában)
correct_prediction = tf.equal(tf.argmax(y,1), tf.argmax(y_,1))
# ez pedig kitszámolja ezek átlagát
accuracy = tf.reduce_mean(tf.cast(correct_prediction, tf.float32))
# most pedig lefuttatjuk a teszt képekre
print(accuracy.eval(session=sess, feed_dict={x: x_test, y_: y_test}))
'''
# + [markdown] id="aE3bC9TVykiM" colab_type="text"
# # convnet
#
# + id="T1TODTrGnQ19" colab_type="code" colab={}
import tensorflow as tf
sess = tf.Session()
def weight_variable(shape):
initial = tf.truncated_normal(shape, stddev = 0.1, mean=0.2, name='weights')
return tf.Variable(initial)
def bias_variable(shape):
initial = tf.constant(0.2, shape=shape)
return tf.Variable(initial)
def conv2d(x, W, name):
return tf.nn.conv2d(x, W, strides=[1,1,1,1], padding='SAME', name=name)
def max_pool_2x2(x):
return tf.nn.max_pool(x, ksize=[1,2,2,1], strides=[1,2,2,1], padding='SAME')
# + id="KCUasp7lnRTU" colab_type="code" colab={}
# bemenet és kimenet definiálása
x = tf.placeholder(tf.float32, shape=[None, x_train.shape[-1]])
y_ = tf.placeholder(tf.float32, shape=[None, y_train.shape[-1]])
# convnet osszerakasa
x_image = tf.reshape(x, [-1,28,28,1])
# 1@28x28, f:5x5@32....
W_conv1 = weight_variable([5,5,1,32])
b_conv1 = bias_variable([32])
h_conv1 = tf.nn.sigmoid(conv2d(x_image, W_conv1, "h_conv1")+b_conv1) # 1@28x28->28x28@32
h_pool1 = max_pool_2x2(h_conv1) # 28x28->14x14
# 32@14x14, f: 5x5@64
W_conv2 = weight_variable([5,5,32,64])
b_conv2 = bias_variable([64])
h_conv2 = tf.nn.relu(conv2d(h_pool1, W_conv2, "h_conv2")+b_conv2)
h_pool2 = max_pool_2x2(h_conv2) # 14x14->7x7 (@64)
# fully connected
W_fc1 = weight_variable([7*7*64,1024])
b_fc1 = bias_variable([1024])
h_flat = tf.reshape(h_pool2, [-1, 7*7*64])
h_fc1 = tf.nn.relu(tf.matmul(h_flat, W_fc1)+b_fc1)
# dropout
keep_prob = tf.placeholder(tf.float32)
h_fc1_drop = tf.nn.dropout(h_fc1, keep_prob)
# kimeneti softmax layer
W_fc2 = weight_variable([1024,10])
b_fc2 = bias_variable([10])
y_conv = tf.nn.softmax(tf.matmul(h_fc1_drop, W_fc2)+b_fc2)
# + id="fB5clwjQnRQS" colab_type="code" colab={}
cross_entropy = tf.reduce_mean(-tf.reduce_sum(y_ * tf.log(y_conv), reduction_indices=[1]))
# optimációs algoritmus megadása
train_step = tf.train.AdamOptimizer(1e-4).minimize(cross_entropy)
correct_prediction = tf.equal(tf.argmax(y_conv,1), tf.argmax(y_,1))
accuracy = tf.reduce_mean(tf.cast(correct_prediction, tf.float32) )
# + id="NnVo84LInROI" colab_type="code" colab={}
sess.run(tf.global_variables_initializer())
batch_counter=0
# + id="S2Ce5fgSnRLw" colab_type="code" outputId="468acfe2-19ab-498b-be81-5d7ed8354c0d" colab={"base_uri": "https://localhost:8080/", "height": 1368}
for i in range(1000):
if i%100==0:
train_acc = accuracy.eval(session=sess, feed_dict={x: x_train[batch_counter*batch_size:(batch_counter+1)*batch_size], y_: y_train[batch_counter*batch_size:(batch_counter+1)*batch_size], keep_prob: 1})
print(i,'step, train accuracy on minibatch: ',train_acc)
train_step.run(session=sess, feed_dict={x: x_train[batch_counter*batch_size:(batch_counter+1)*batch_size], y_: y_train[batch_counter*batch_size:(batch_counter+1)*batch_size], keep_prob: 0.5})
batch_counter=batch_counter+1
if (batch_counter*batch_size > len(x_train)):
batch_counter=0
print("test acc: %f" % accuracy.eval(session=sess, feed_dict={x: x_test, y_: y_test, keep_prob: 1}))
# + id="6Gr72GGWnRJQ" colab_type="code" colab={}
# + id="q9zmgRG6nRGb" colab_type="code" colab={}
# + id="t1zydOeknRDZ" colab_type="code" colab={}
# + id="zIfTQ7nAnQ0G" colab_type="code" colab={}
# + id="pCdgwVXfnQxp" colab_type="code" colab={}
# + id="Mz-pb8h0nQvA" colab_type="code" colab={}
# + id="mDgFE16unQsC" colab_type="code" colab={}
# + [markdown] id="aT2lZ0mXl38G" colab_type="text"
# # Regebbiek
# + id="hQ3L_DuGirdd" colab_type="code" colab={}
import numpy as np
np.random.seed(42)
# + id="DIy0_TkgiwOF" colab_type="code" outputId="46ca28ce-ca1e-4ab1-c376-a23ef1df64f7" colab={"base_uri": "https://localhost:8080/", "height": 105}
np.random.rand(5,3)
# + id="rQ0NtwZ0jLTg" colab_type="code" outputId="7589ad28-664a-4534-b348-a6ca3a9f2020" colab={"base_uri": "https://localhost:8080/", "height": 105}
np.random.rand(5,3)
# + id="KG1P6-vHjM4E" colab_type="code" colab={}
from tensorflow import set_random_seed
set_random_seed(123)
# + id="qN5wMlgijVBf" colab_type="code" outputId="2073e99a-2890-4206-b4dc-b903486d9427" colab={"base_uri": "https://localhost:8080/", "height": 35}
from keras.models import Sequential
from keras.layers.core import Dense, Activation
from keras.callbacks import Callback
from keras.optimizers import SGD
from sklearn.preprocessing import StandardScaler, MinMaxScaler
# %matplotlib inline
import matplotlib.pyplot as plt
# + id="NFShzUt6j54X" colab_type="code" colab={}
nb_samples = 2000
valid_split = 0.2
test_split = 0.1
samples = np.zeros(nb_samples, \
dtype=[('input', float, 2), ('output', float, 1)])
for i in range(0,nb_samples,4):
noise = np.random.normal(0,1,8)
samples[i] = (-2+noise[0], -2+noise[1]), 0
samples[i+1] = (2+noise[2], -2+noise[3]), 1
samples[i+2] = (-2+noise[4], 2+noise[5]), 1
samples[i+3] = (2+noise[6], 2+noise[7]), 0
# + id="opWFQw9Ck1Ic" colab_type="code" outputId="b22911c9-9b0d-435e-cce9-75078cf5b4aa" colab={"base_uri": "https://localhost:8080/", "height": 283}
fig1 = plt.figure()
plt.scatter(samples['input'][:,0], samples['input'][:,1], \
c=samples['output'][:])
# + id="_xHxihtsk_0u" colab_type="code" colab={}
# train-valid-test split #### valid_split, test_split
samples_train = samples[:int(nb_samples*(1-valid_split-test_split))]
samples_valid = samples[int(nb_samples*(1-valid_split-test_split)):int(nb_samples*(1-test_split))]
samples_test = samples[int(nb_samples*(1-test_split)):]
# + id="UCE-Los5oGVs" colab_type="code" outputId="5fabca29-d302-438d-94d0-f6510166ef21" colab={"base_uri": "https://localhost:8080/", "height": 35}
len(samples_train), len(samples_valid), len(samples_test)
# + id="L73EV5cwmEyw" colab_type="code" colab={}
# standardizalas train alapjan train, valid es test-re
scaler = StandardScaler().fit(samples_train['input']) # mean es var kiszamolasa
samples_train['input'] = scaler.transform(samples_train['input'] ) # (argargarg-mean)/stdev
samples_valid['input'] = scaler.transform(samples_valid['input'])
samples_test['input'] = scaler.transform(samples_test['input'])
# + id="8WCpQq_HmGDi" colab_type="code" outputId="b8705bec-51a1-4455-f754-eb3739a58c55" colab={"base_uri": "https://localhost:8080/", "height": 35}
np.mean(samples_train['input']), np.std(samples_train['input'])
# + id="5bCNyqgbmIBm" colab_type="code" outputId="386bedf0-1447-467c-8f75-cbf4a1f3e0c4" colab={"base_uri": "https://localhost:8080/", "height": 35}
np.mean(samples_valid['input']), np.std(samples_valid['input'])
# + id="rEdOgSsWovPH" colab_type="code" outputId="1c4c79c8-7b48-4bf5-b9f5-a0ec33ce1a9c" colab={"base_uri": "https://localhost:8080/", "height": 34}
np.mean(samples_test['input']), np.std(samples_test['input'])
# + id="T9RvPQ_Ao0SE" colab_type="code" outputId="6050abd5-2bcc-4751-e1aa-45f963bc64a3" colab={"base_uri": "https://localhost:8080/", "height": 282}
fig1 = plt.figure()
plt.scatter(samples_train['input'][:,0], samples_train['input'][:,1], \
c=samples_train['output'][:])
# + id="4f9sodW0vx8W" colab_type="code" colab={}
class TrainingHistory(Callback):
def on_train_begin(self, logs={}):
self.losses=[]
self.valid_losses =[]
self.accs = []
self.valid_accs = []
self.epoch=0
def on_epoch_end(self, epoch,logs={}):
self.losses.append(logs.get('loss'))
self.valid_losses.append(logs.get('val_loss'))
self.accs.append(logs.get('acc'))
self.valid_accs.append(logs.get('val_acc'))
self.epoch += 1
history = TrainingHistory()
# + id="FD78NMy90XNl" colab_type="code" colab={}
from keras.callbacks import EarlyStopping
es = EarlyStopping(patience=10, verbose=1)
from keras.callbacks import ModelCheckpoint
mcp = ModelCheckpoint(filepath='weights.hdf5', verbose=1, save_best_only=True)
# + id="C8VpG_popCuB" colab_type="code" colab={}
from keras import regularizers
# + id="JOOy4RYXpAWc" colab_type="code" colab={}
# Neuralis halo felepites
model = Sequential()
model.add(Dense(100, input_shape=(2,), use_bias=True, kernel_regularizer=regularizers.l2(0.01)))
model.add(Activation('tanh'))
model.add(Dense(100, activation='relu'))
model.add(Dense(1, use_bias=False))
model.add(Activation('sigmoid'))
# + id="sEDpVbtupwXB" colab_type="code" outputId="630a13b0-6396-498c-c619-45b51c58ee92" colab={"base_uri": "https://localhost:8080/", "height": 4024}
sgd=SGD(lr=0.1, momentum=0.001, nesterov=True)
model.compile(loss='mean_squared_error', optimizer=sgd)
hst = model.fit(samples_train['input'], samples_train['output'],\
batch_size=8,
epochs=2500000000,
verbose=2,
validation_data=(samples_valid['input'],samples_valid['output']),
callbacks=[mcp, es, history],
shuffle=True)
# + id="1GyEwlShqpcq" colab_type="code" outputId="4e0ed6fa-0f0b-4685-ada8-47841672a32c" colab={"base_uri": "https://localhost:8080/", "height": 241}
plt.figure(figsize=(6,3))
plt.title("Hiba mértéke a tanítás során")
plt.plot(np.arange(history.epoch), history.losses, color='g',
label="Hiba a tanító adatokon")
plt.plot(np.arange(history.epoch), history.valid_losses, color='r',
label="Hiba a validációs adatokon")
plt.legend(loc='upper right')
plt.xlabel("Epochok száma")
plt.ylabel("Hiba")
plt.grid(True)
plt.show()
# + id="aPfRS7bZz4hf" colab_type="code" colab={}
from keras.models import load_model
model = load_model('weights.hdf5')
# + id="hUtmfuN_2sie" colab_type="code" outputId="78a57e98-f953-4015-af6c-9f9befcec9fd" colab={"base_uri": "https://localhost:8080/", "height": 3484}
preds = model.predict(samples_test['input'])
print(preds)
# + id="2HVwS6RT2yKb" colab_type="code" outputId="2b47a02b-ded8-42a0-8fb5-ef5a4481d5e7" colab={"base_uri": "https://localhost:8080/", "height": 35}
from sklearn.metrics import mean_squared_error
test_mse = mean_squared_error(samples_test['output'], preds)
print("Teszt adatokon mért négyzetes hiba:",test_mse)
# + id="kgaPOdzV3IQJ" colab_type="code" outputId="e08e4b05-8fc7-487f-8743-e326b9aaa859" colab={"base_uri": "https://localhost:8080/", "height": 35}
print("Tehát átlagosan",np.sqrt(test_mse),"hibázik a hálónk a teszt adatokon")
# + id="mlPdojfv3QUV" colab_type="code" outputId="3518b8c4-b0aa-4f28-9908-25a0d36a1159" colab={"base_uri": "https://localhost:8080/", "height": 283}
plt.figure()
plt.scatter(samples_test['input'][:,0], samples_test['input'][:,1],
c=np.round(preds[:,0]))
# + id="6lfYWW-O3lhV" colab_type="code" colab={}
# + [markdown] id="BqO9Y9jN3war" colab_type="text"
# # Modell vizsgálata
# + id="mnFn2RK63xis" colab_type="code" outputId="aeb08c63-6e6a-4609-ab97-d2861bb866cf" colab={"base_uri": "https://localhost:8080/", "height": 312}
model.summary()
# + id="eq5_efGe3zQa" colab_type="code" outputId="2a4e6135-6629-4cbf-ac7d-e191f6ca1827" colab={"base_uri": "https://localhost:8080/", "height": 228}
print("Rétegek")
for layer in model.layers:
print("Réteg neve:", layer.name,", tanítható: ", layer.trainable)
print(layer.get_config())
# + id="_CvUrI6B4oT5" colab_type="code" outputId="f3e4c5fc-7b58-497b-9ab3-a43792589718" colab={"base_uri": "https://localhost:8080/", "height": 3920}
print("Súlyok:")
i=1
for layer in model.layers:
print(model.get_layer(index=i).get_weights())
i+=1
# + id="B47Vfhww4EPr" colab_type="code" colab={}
w_l1 = model.get_layer(index=1).get_weights()
# + id="PKd4C4qzskaX" colab_type="code" outputId="e9c44a1d-d9b0-4645-b05f-e43b8f18e0e0" colab={"base_uri": "https://localhost:8080/", "height": 433}
seged = w_l1[0][0]
print(seged[np.abs(seged)<0.001])
# + id="iXk0hANHsk51" colab_type="code" colab={}
# + [markdown] id="aUy4f7q8yvYI" colab_type="text"
# # Boston Housing Prices
# + id="8ZRg4-EgyxQ2" colab_type="code" outputId="97580d12-453b-47c7-a50c-fe98b8f919af" colab={"base_uri": "https://localhost:8080/", "height": 228}
# !wget https://archive.ics.uci.edu/ml/machine-learning-databases/housing/housing.data
# + id="lyq7dofq0Ksq" colab_type="code" outputId="1ef29f8f-7f7a-46cc-e7bc-0d34a3242400" colab={"base_uri": "https://localhost:8080/", "height": 35}
# !ls
# + id="ZQnHxyEl0MUF" colab_type="code" colab={}
import numpy as np
from keras.models import Sequential
from keras.layers import Dense, Dropout, Activation
from keras.callbacks import EarlyStopping, ModelCheckpoint
import copy
import pandas as pd
from keras.optimizers import SGD
from sklearn.preprocessing import StandardScaler
from tensorflow import set_random_seed
set_random_seed(123)
np.random.seed(123)
# + id="bgpolaGp0ixW" colab_type="code" colab={}
df = pd.read_csv("housing.data", delim_whitespace=True, header=None)
dataset=df.values
# + id="8l_P_d7O00mJ" colab_type="code" outputId="0ebbf55a-c79d-4fd3-9fb7-e84c0a71982f" colab={"base_uri": "https://localhost:8080/", "height": 35}
dataset.shape
# + id="XKYLwl6602Rc" colab_type="code" colab={}
test_split = 0.1
valid_split = 0.1
X = dataset[:,:13]
Y = dataset[:,13]
v_index = int(X.shape[0]*(1-valid_split-test_split))
t_index = int(X.shape[0]*(1-test_split))
X_test = X[t_index:]
Y_test = Y[t_index:]
X_valid = X[v_index:t_index]
Y_valid = Y[v_index:t_index]
X_train = X[:v_index]
Y_train = Y[:v_index]
scaler = StandardScaler().fit(X_train)
X_train = scaler.transform(X_train)
X_valid = scaler.transform(X_valid)
X_test = scaler.transform(X_test)
# + id="BiuIIqut1uMa" colab_type="code" outputId="2c20fd86-13b9-4090-c508-4b2530b5d90c" colab={"base_uri": "https://localhost:8080/", "height": 2291}
es = EarlyStopping(patience = 30)
mcp = ModelCheckpoint(filepath='weights.hdf5', verbose=1, save_best_only=True)
model = Sequential()
model.add(Dense(60, input_shape=(X_train.shape[1],)))
model.add(Activation('sigmoid'))
model.add(Dropout(0.3))
model.add(Dense(1, activation='linear'))
sgd = SGD(lr=1e-3, decay=1e-6, momentum=0.9, nesterov=True)
model.compile(loss='mse', optimizer=sgd)
history = model.fit(X_train, Y_train,
epochs=10000000,
batch_size=8,
verbose=2,
validation_data=(X_valid,Y_valid),
callbacks=[mcp,es])
# + id="o1vGYW7i22kU" colab_type="code" outputId="52f0c1ca-2cf0-4107-e824-4c2a93fad5ba" colab={"base_uri": "https://localhost:8080/", "height": 176}
# !pip install keras --upgrade
# + id="WGxW73dO3oRk" colab_type="code" outputId="8b3f3c98-2cc2-4daa-c450-6b4f9cceaf59" colab={"base_uri": "https://localhost:8080/", "height": 314}
df.describe()
# + id="fl8DV_Uz27cs" colab_type="code" colab={}
from keras.models import load_model
model = load_model('weights.hdf5')
# + id="JpUfKDIz4KMv" colab_type="code" outputId="0943d75a-f0cf-4ed8-e116-2c0b861d9b7c" colab={"base_uri": "https://localhost:8080/", "height": 35}
from sklearn.metrics import mean_squared_error
preds = model.predict(X_test)
err = mean_squared_error(Y_test,preds)
print("teszt adatokon mert hiba:",err)
# + id="0lNuQBdX4VuO" colab_type="code" outputId="376f26ef-7ea4-4f6e-da6c-2baeecc25f87" colab={"base_uri": "https://localhost:8080/", "height": 35}
np.sqrt(err)
# + id="0FaZ68J04Y8M" colab_type="code" outputId="e5e1528d-d2d0-478b-9da6-48b8f73d8f74" colab={"base_uri": "https://localhost:8080/", "height": 615}
# %matplotlib inline
import seaborn as sns # matplotlib, bokeh, ....
import matplotlib.pyplot as plt
plt.figure(figsize=(10,10))
sns.regplot(x=Y_test, y=preds.reshape(-1)).set(xlim=(10,30),ylim=(10,30))
# + id="QfaPYxdD4-c3" colab_type="code" colab={}
preds_mean = [np.mean(Y_train) for i in range(Y_test.shape[0])]
# + id="GjqbSNCJ5hRg" colab_type="code" outputId="bb25d6db-488a-4e17-bc2d-4325d1edc290" colab={"base_uri": "https://localhost:8080/", "height": 55}
print(preds_mean)
# + id="8MZ007D35zc8" colab_type="code" colab={}
preds_mean = np.array(preds_mean)
# + id="GGa7rluI5j8d" colab_type="code" outputId="ac7a4d6d-1cf4-4c5e-90f1-200f7f3447be" colab={"base_uri": "https://localhost:8080/", "height": 35}
Y_test.shape
# + id="JIZfA2Z_5l14" colab_type="code" outputId="2f14205c-05e9-419f-af69-832b60d9c994" colab={"base_uri": "https://localhost:8080/", "height": 35}
from sklearn.metrics import mean_squared_error
err_mean = mean_squared_error(Y_test,preds_mean)
print("teszt adatokon mert hiba:",err_mean)
# + id="L1ZmWGKS55lQ" colab_type="code" outputId="cea9b457-7e51-4caf-99b5-2bfeeabd0531" colab={"base_uri": "https://localhost:8080/", "height": 615}
# %matplotlib inline
import seaborn as sns # matplotlib, bokeh, ....
import matplotlib.pyplot as plt
plt.figure(figsize=(10,10))
sns.regplot(x=Y_test, y=preds_mean.reshape(-1)).set(xlim=(10,30),ylim=(10,30))
# + id="j-inHVsB59Lh" colab_type="code" colab={}
| 30,814 |
/LectureNotes/ch00python/030MazeSolution.ipynb | ab5dac5cdb3a6c4a8a61062f4bc161aa744fce08 | [
"CC-BY-4.0",
"CC-BY-3.0"
] | permissive | annaschroder/exam_prep | https://github.com/annaschroder/exam_prep | 0 | 0 | null | null | null | null | Jupyter Notebook | false | false | .py | 2,709 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# ### Solution: my Maze Model
# Here's one possible solution to the Maze model. Yours will probably be different, and might be just as good.
# That's the artistry of software engineering: some solutions will be faster, others use less memory, while others will
# be easier for other people to understand. Optimising and balancing these factors is fun!
house = {
'living' : {
'exits': {
'north' : 'kitchen',
'outside' : 'garden',
'upstairs' : 'bedroom'
},
'people' : ['James'],
'capacity' : 2
},
'kitchen' : {
'exits': {
'south' : 'living'
},
'people' : [],
'capacity' : 1
},
'garden' : {
'exits': {
'inside' : 'living'
},
'people' : ['Sue'],
'capacity' : 3
},
'bedroom' : {
'exits': {
'downstairs' : 'living',
'jump' : 'garden'
},
'people' : [],
'capacity' : 1
}
}
# Some important points:
# * The whole solution is a complete nested structure.
# * I used indenting to make the structure easier to read.
# * Python allows code to continue over multiple lines, so long as sets of brackets are not finished.
# * There is an **empty** person list in empty rooms, so the type structure is robust to potential movements of people.
# * We are nesting dictionaries and lists, with string and integer data.
| 1,713 |
/diabetes_background_research.ipynb | 815600d9607729e44cfd207cec49ef37a724d32b | [] | no_license | akshay43279/background_research | https://github.com/akshay43279/background_research | 0 | 0 | null | null | null | null | Jupyter Notebook | false | false | .py | 534,345 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# %matplotlib inline
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import warnings
warnings.filterwarnings('ignore')
diabetes = pd.read_csv('diabetes.csv')
diabetes.head()
diabetes.shape
diabetes.columns
# ### EDA for the dataset has been done using Tableau
# ### Missing or Null values
diabetes.isnull().sum()
diabetes.isna().sum()
# #### No missing or null values are present in the dataset
# ### Outlier detection and treatment
diabetes[diabetes.BloodPressure == 0].shape[0]
# ##### A living patient cannot have a blood pressure of zero.
diabetes[diabetes.Glucose == 0].shape[0]
# ##### Glucose level can never be equal to zero.
diabetes[diabetes.SkinThickness == 0].shape[0]
# ##### Skin Fold thickness can be rarely less than 10mm. Zero is not remotely possible.
diabetes[diabetes.BMI == 0].shape[0]
# ##### BMI can never be zero even if the patient is very very underweight.
diabetes[diabetes.Insulin == 0].shape[0]
# ##### In a very rare situation, a person can have zero insulin.
# #### All the above observations show that there have been some discrepancies in the data input process or that simply the data was not available for all the patient records.
#
# #### We will drop the rows where Blood Pressure, BMI and Glucose have zero values.
diabetes_after_dc = diabetes[(diabetes.BloodPressure != 0) & (diabetes.BMI != 0) & (diabetes.Glucose != 0)]
diabetes_after_dc.shape
diabetes_after_dc.columns
# #### Separating the dataset into predictor and response variables
features = ['Pregnancies', 'Glucose', 'BloodPressure', 'SkinThickness', 'Insulin', 'BMI', 'DiabetesPedigreeFunction', 'Age']
X = diabetes_after_dc[features]
y = diabetes_after_dc.Outcome
X.head()
y.head()
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size = 0.25, stratify = diabetes_after_dc.Outcome, random_state=0)
# ### Random Forest Model
from sklearn.ensemble import RandomForestClassifier
rf = RandomForestClassifier(n_estimators=100, random_state=0)
rf.fit(X_train, y_train)
print("Accuracy on training set: {:.3f}".format(rf.score(X_train, y_train)))
print("Accuracy on test set: {:.3f}".format(rf.score(X_test, y_test)))
from sklearn import metrics
feature_names = X_train.columns
feature_imports = rf.feature_importances_
most_imp_features = pd.DataFrame([f for f in zip(feature_names,feature_imports)], columns=["Feature", "Importance"]).nlargest(10, "Importance")
most_imp_features.sort_values(by="Importance", inplace=True)
plt.figure(figsize=(10,6))
plt.barh(range(len(most_imp_features)), most_imp_features.Importance, align='center', alpha=0.8)
plt.yticks(range(len(most_imp_features)), most_imp_features.Feature, fontsize=14)
plt.xlabel('Importance')
plt.title('Most important features - Random Forest')
plt.show()
import shap # package to calculate SHAP values
X_test.head()
row_to_show = 0
data_for_prediction = X_test.iloc[row_to_show]
data_for_prediction_array = data_for_prediction.values.reshape(1, -1)
rf.predict_proba(data_for_prediction_array)
# Creating an object to explain SHAP values
explainer = shap.TreeExplainer(rf)
shap_values = explainer.shap_values(data_for_prediction)
print(shap_values)
shap.initjs()
shap.force_plot(explainer.expected_value[1], shap_values[1], data_for_prediction)
explainer = shap.TreeExplainer(rf)
shap_values = explainer.shap_values(X_test)
shap.summary_plot(shap_values[1], X_test)
shap.dependence_plot('Glucose', shap_values[1], X_test, interaction_index = "BMI")
| 3,837 |
/notebooks/ak1982/netflix-tv-show-and-movie-analyiss.ipynb | b7cdff78399f6eab139cbc114e3929af20c783bc | [] | no_license | Sayem-Mohammad-Imtiaz/kaggle-notebooks | https://github.com/Sayem-Mohammad-Imtiaz/kaggle-notebooks | 5 | 6 | null | null | null | null | Jupyter Notebook | false | false | .py | 68,744 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# <h1><center>Netflix TV Shows And Movie Analysis</center></h1>
#
# 
#
#
# ##### Image credit:https://streamdiag.com/netflix-is-stuck-on-loading-screen-fix/
# Netflix was founded in 1997 by Reed Hastings and Marc Randolph
# The company's primary business is a subscription-based streaming service offering online streaming.
# Netflix is headquartered in Los Gatos, California.
# In this Analysis we will answer the following questions through visualistions
#
# ### Questions
# 1. What contents Neflix has most
# 2. Top contents by ratings
# 3. Countries with most content
# 4. Busiest month for content release
# 5. Total contents by Season
# 6. Top ten TV shows
# 7. Movie released by ratings since 2016
# 8. Tv Shows India has by ratings
# 9. Tv Shows USA has by ratings
# 10. Top 10 Movie genre
# ### Import Necessary Libraries
# + _uuid="8f2839f25d086af736a60e9eeb907d3b93b6e0e5" _cell_guid="b1076dfc-b9ad-4769-8c92-a6c4dae69d19" _kg_hide-input=true
import pandas as pd
import os
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
from wordcloud import WordCloud, STOPWORDS, ImageColorGenerator
from PIL import Image
# %matplotlib inline
import requests
import re
# -
# ### Load the Dataset
netflix = pd.read_csv('../input/netflix-shows/netflix_titles.csv')
netflix.head(3)
# # Exploratory Data Analysis
netflix.info()
netflix.columns
netflix.nunique()
# ### Check for Missing Values
# + _kg_hide-input=true
def missing_value_table(df):
missing_value = df.isna().sum().sort_values(ascending=False)
missing_value_percent = 100 * df.isna().sum()//len(df)
missing_value_table = pd.concat([missing_value, missing_value_percent], axis=1)
missing_value_table_return = missing_value_table.rename(columns = {0 : 'Missing Values', 1 : '% Value'})
cm = sns.light_palette("green", as_cmap=True)
missing_value_table_return = missing_value_table_return.style.background_gradient(cmap=cm)
return missing_value_table_return
missing_value_table(netflix)
# -
# ### Handling Missing Values
#
# Replace missing values with mode for ratings and country column
# + _kg_hide-input=false
netflix['rating'] = netflix['rating'].fillna(netflix['rating'].mode()[0])
netflix['country'] = netflix['country'].fillna(netflix['country'].mode()[0])
netflix[['rating','country']].isna().sum()
# -
# Replace nan values in data_added with January 1
netflix['date_added']=netflix['date_added'].fillna('January 1, {}'.format(str(netflix['release_year'].mode()[0])))
netflix['date_added'].isna().sum()
# Drop missing value rows from director and cast columns
netflix.drop(['director','cast'],axis = 1,inplace = True)
# verify dataset has no missing values
netflix.isna().sum()
# Convert duration to numeric
netflix['duration'] = netflix['duration'].map(lambda x : re.sub('[^0-9]','',x))
netflix['duration'] = pd.to_numeric(netflix['duration'])
netflix.head(3)
# # Data visualization
#
# ##### Movies vs TV Show
# + _kg_hide-input=true
plt.figure(figsize= (8,5))
sns.set_style(style = 'darkgrid')
ax = sns.countplot(x = 'type',data = netflix, palette = 'Set2')
plt.title('Movie Vs TV Show')
# -
# ##### Number of Contents by Ratings
# + _kg_hide-input=true
sns.set_style(style = 'darkgrid')
plt.figure(figsize = (10,3))
ax = sns.countplot(x ='rating', data = netflix,palette = 'Set2')
plt.title('Count of Content Ratings',fontsize = 15)
plt.xticks(rotation =90)
plt.xlabel('Ratings',fontsize = 15)
plt.ylabel('Total Count',fontsize = 15)
# -
# ##### Total number of Tv Shows and Movies by Ratings
# + _kg_hide-input=true
sns.set_style(style = 'darkgrid')
plt.figure(figsize = (10,8))
ax = sns.countplot(data = netflix, x= 'rating',hue = 'type', palette='Set2' )
plt.xticks(rotation = 90)
plt.title('Content Ratings TV Shows vs Movie', fontsize = 15)
plt.xlabel('Ratings',fontsize = 15)
plt.ylabel('Total Count',fontsize = 15)
# -
# ##### Contries with most contents
# + _kg_hide-input=true
plt.figure(figsize = (10,5))
ax = sns.countplot(x = 'country',data= netflix, palette ='gist_rainbow', order = netflix['country'].value_counts().index[0:15])
plt.xlabel('Country')
plt.xticks(rotation = 90)
plt.title('Countries with most content(Top 15)')
plt.ylabel('Number of Movies and TV Shows')
# -
# ##### Busiest Month for Content Release
# + _kg_hide-input=true
netflix['release_month'] = netflix['date_added'].apply(lambda x: x.lstrip().split(' ')[0])
sns.countplot(y = 'release_month',data = netflix,palette ='Pastel1', order = netflix['release_month'].value_counts().index)
plt.title('Movies/Tv show releases by month')
plt.ylabel('Release Month')
plt.xlabel('No. of Releases')
# -
# ##### Count of TV Shows by number of seasons
# + _kg_hide-input=true
plt.figure(figsize= (10,5))
tv_show = netflix[netflix['type']== 'TV Show']
sns.countplot(x = 'duration',data = tv_show,palette = 'Oranges_r', order = tv_show['duration'].value_counts().index)
plt.xticks(rotation = 90)
plt.xlabel("Seasons",fontsize = 15)
plt.ylabel("Total count",fontsize = 15)
plt.title("Total Tv Show Season wise",fontsize = 15)
# -
# ##### Movie Duration
# + _kg_hide-input=true
movie_duration = netflix.loc[netflix['type']=='Movie']
plt.figure(figsize=(12,10))
ax = sns.histplot(data = movie_duration,x = 'duration',bins = 50,kde = True,color = 'red')
plt.title('Movie Duration',fontsize = 15)
plt.xlabel('Total Duration(In Mins)',fontsize = 15)
plt.ylabel('Total Movie Count',fontsize = 15)
# -
# ##### TOP 10 TV Show
# + _kg_hide-input=true
tv_shows = netflix.loc[netflix['type']=='TV Show']
top10_shows = tv_shows.sort_values(by=['duration'])[-1:-11:-1]
top10_shows[['title','duration']]
plt.figure(figsize=(8,10))
sns.barplot(y = top10_shows['title'], x = top10_shows['duration'])
plt.title('Top 10 TV Shows',fontsize = 15)
plt.xlabel('No.of Seasons',fontsize = 15)
plt.ylabel('TV Shows',fontsize = 15)
# -
# ##### Trend of movie ratings that has been released over the last five years
#
# + _kg_hide-input=true
movie = netflix.loc[netflix['type']=='Movie']
movie_trends = movie[movie['release_year']>=2016]
df = movie_trends.groupby(['release_year','rating']).size().reset_index(name = 'Total')
df1 = df[df['Total'] >= 10]
plt.figure(figsize = (10,8))
sns.set_style('darkgrid')
sns.barplot(data = df1,x = 'release_year', y = 'Total',hue = 'rating',palette = 'Set1')
plt.title('Movies released by rating since 2016 and beyond(more than 10 releases per year)')
plt.xlabel('Year')
plt.ylabel('Total Count')
# -
# ##### Tv Shows India has by rating
# + _kg_hide-input=true
df = netflix[(netflix['type'] == 'TV Show') & (netflix['country'] == 'India') ]
tv_show_India = df.groupby(['release_year','rating']).size().reset_index(name = 'Total')
plt.figure(figsize = (12,10))
sns.set_style('darkgrid')
sns.barplot(data = tv_show_India,x = 'release_year', y = 'Total',hue = 'rating',palette = 'Set1')
plt.title('TV shows released by India by ratings',fontsize = 15)
plt.xlabel('Year',fontsize = 15)
plt.ylabel('No.of Shows',fontsize = 15)
# -
# ##### Tv Shows USA has by rating
# + _kg_hide-input=true
tv_show_USA= netflix[(netflix['type'] == 'TV Show') & (netflix['country'] == 'United States') ]
tv_show_USA = tv_show_USA[tv_show_USA['release_year']>=2010]
df = tv_show_USA.groupby(['release_year','rating']).size().reset_index(name = 'Total')
plt.figure(figsize = (12,10))
sns.set_style('darkgrid')
sns.barplot(data = df,x = 'release_year', y = 'Total',hue = 'rating',palette = 'Set1')
plt.title('TV shows released by United States by ratings')
# -
# ##### Top Ten Movie genre
# + _kg_hide-input=true
movie_genre = netflix.loc[netflix['type'] == 'Movie']
plt.figure(figsize=(12,6))
ax = sns.countplot(y='listed_in',data = movie_genre,order = movie_genre["listed_in"].value_counts().index[0:10],palette="Set2")
plt.title('Top Ten Movie Genre',fontsize = 15)
plt.xlabel('Total Count',fontsize = 15)
plt.ylabel('Genre',fontsize = 15)
# -
# ##### Top Ten TV Show genre
# + _kg_hide-input=true
Tv_Show_genre = netflix.loc[netflix['type'] == 'TV Show']
plt.figure(figsize=(12,6))
ax = sns.countplot(y='listed_in',data = Tv_Show_genre,order = Tv_Show_genre["listed_in"].value_counts().index[0:10],palette="Set2")
plt.title('Top Ten Tv Show Genre',fontsize = 15)
plt.xlabel('Total Count',fontsize = 15)
plt.ylabel('Genre',fontsize = 15)
# -
# ### Trends of content released Year over Year
# + _kg_hide-input=true
df = netflix[netflix['type'] == 'TV Show'].groupby('release_year').count()[-25:-1]
df1 = netflix[netflix['type'] == 'Movie'].groupby('release_year').count()[-25:-1]
df2 = netflix.groupby('release_year').count()[-25:-1]
plt.figure(figsize = (15,8))
sns.set_style('darkgrid')
sns.lineplot(data = df['show_id'],palette = 'Set1')
sns.lineplot(data = df1['show_id'],palette = 'Set1')
sns.lineplot(data = df2['show_id'],palette = 'Set1')
plt.title('Year over Year Content Released',fontsize = 15)
plt.xlabel('Year',fontsize = 15)
plt.ylabel('No.of Shows',fontsize = 15)
plt.legend(['TV', 'Movie','Total'], fontsize='large')
# -
# ### Word Cloud
#
# ##### Count of most used words in 'listed in'
# + _kg_hide-input=true
stopwords = set(STOPWORDS)
wordcloud = WordCloud(background_color='white',
stopwords=stopwords,
max_words=200,
max_font_size=40,
random_state=42
).generate(str(netflix['listed_in']))
plt.imshow(wordcloud)
plt.axis('off')
plt.show()
# -
# ##### Count of most used word in 'Description'
# + _kg_hide-input=true
words = (str(netflix['description']))
mask = np.array(Image.open(requests.get('https://i.stack.imgur.com/B0AAW.png',stream=True).raw))
def create_wordcloud(words, mask):
wc = WordCloud(width =500, height =800, background_color='white',
stopwords=STOPWORDS, mask=mask,max_font_size =40,
repeat=True,max_words = 300).generate(words)
plt.figure(figsize=(15,15))
plt.imshow(wc)
plt.axis('off')
plt.show()
create_wordcloud(words, mask)
# -
# ### Findings
# 1. Netflix has more movies than Tv shows
# 2. Netflix has TV-MA ratings the most
# 3. USA is by far the top contributer followed by India
# 4. December is the busiest month for contents release followed by October
# 5. Most of the contents has only 1 season
# 6. Grey's Anatomy tops the list with most seasons(16) followed by NCIS. Cheers is number 10 with 10 seasons
# 7. Since 2016 TV-MA has been released the most
# 8. India has shifted towards TV-MA ratings after 2017. Before that it was mostly TV-14 ratings
# 9. Same with USA, TV-MA content has increased since 2017.
| 10,940 |
/tensorflow单层神经网络(MNIST手写数字识别进阶1).ipynb | 79c68cd46d97e2d80410162b86324203e0e8d437 | [] | no_license | zhangziqi1999/tensorflow-notebook | https://github.com/zhangziqi1999/tensorflow-notebook | 0 | 0 | null | null | null | null | Jupyter Notebook | false | false | .py | 704,006 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# 
# # 常见激活函数
# 
# # 全连接单隐藏层神经网络结构图
# 
# # 全连接单隐藏层神经网络实现
# 在神经网络中,隐藏层的作用主要是提取数据的特征(feature)。
# # 载入数据:
import tensorflow as tf
import tensorflow.examples.tutorials.mnist.input_data as input_data
#导入tensorflow提供的MNIST模块
mnist = input_data.read_data_sets("MNIST_data/", one_hot = True)
# # 构建输入层
x = tf.placeholder(tf.float32, [None, 784], name = "x")
#mnist中每张图片共有28*28=784个像素点
y = tf.placeholder(tf.float32, [None, 10], name = "y")
#10个数字 = 10个类别
# # 构建隐藏层
# +
H1_NN = 256
#隐藏层神经元数量为256
w1 = tf.Variable(tf.random_normal([784, H1_NN]))
b1 = tf.Variable(tf.zeros([H1_NN]))
y1 = tf.nn.relu(tf.matmul(x, w1) + b1)
# -
# # 构建输出层
# +
w2 = tf.Variable(tf.random_normal([H1_NN, 10]))
b2 = tf.Variable(tf.zeros([10]))
forward = tf.matmul(y1, w2) + b2
pred = tf.nn.softmax(forward)
# -
# # 定义损失函数
loss_function = tf.reduce_mean(tf.nn.softmax_cross_entropy_with_logits(logits = forward, labels = y))
#tensorflow提供了 softmax_cross_entropy_with_logits函数
#用于避免因为log(0)值为NaN造成的数据不稳定
# # 设置训练参数
train_epochs = 40
batch_size = 50
total_batch = int(mnist.train.num_examples / batch_size)
display_step = 1
learning_rate = 0.01
# # 选择优化器
optimizer = tf.train.AdamOptimizer(learning_rate).minimize(loss_function)
# # 定义准确率
correct_prediction = tf.equal(tf.argmax(y, 1), tf.argmax(pred, 1))
accuracy = tf.reduce_mean(tf.cast(correct_prediction, tf.float32))
# # 训练模型
# +
from time import time
starttime = time()
sess = tf.Session()
sess.run(tf.global_variables_initializer())
for epoch in range(train_epochs):
for batch in range(total_batch):
xs, ys = mnist.train.next_batch(batch_size)
#读取批次数据
sess.run(optimizer, feed_dict = {x:xs, y:ys})
#执行批次训练
loss, acc = sess.run([loss_function, accuracy], feed_dict = {x:mnist.validation.images, y:mnist.validation.labels})
#total_batch个批次训练完成后,使用验证数据计算误差和准确率,验证集不分批
if (epoch + 1) % display_step == 0:
print("train epoch:", epoch + 1, "loss =", loss, "accuracy =", acc)
#打印训练详细信息
duration = time() - starttime
print("Train Finished!")
print("It takes", duration, "seconds")
| 2,454 |
/dog_app.ipynb | 301839d919f8067a33838cc8f7183a9e9a5ee6c4 | [] | no_license | bmayank/dog_breed | https://github.com/bmayank/dog_breed | 0 | 0 | null | null | null | null | Jupyter Notebook | false | false | .py | 1,492,617 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# +
from keras.models import Sequential
from keras.layers import Dense
import numpy as np
seed = 7
np.random.seed(seed)
# -
dataset = np.loadtxt("./pimaindiansdiabetescsv/pima-indians-diabetes.csv", delimiter=",")
X = dataset[:,0:8]
Y = dataset[:,8]
X
# +
# create model
model = Sequential()
# 1st layer
model.add(Dense(12, input_dim=8, init='uniform', activation='relu'))
# 2nd layer
model.add(Dense(8, init='uniform', activation='relu'))
# ouput layer
model.add(Dense(1, init='uniform', activation='sigmoid'))
# Compile model
model.compile(loss='binary_crossentropy', optimizer='adam', metrics=['accuracy'])
# Fit the model
model.fit(X, Y, nb_epoch = 150, batch_size=10)
# evaluate the model
score = model.evaluate(X, Y)
print("%s: %.2f%%" %(model.metrics_names[1], score[1]*100))
# -
MEBQYHCAkKC//EALURAAIBAgQEAwQHBQQEAAECdwABAgMRBAUhMQYSQVEHYXETIjKBCBRCkaGxwQkjM1LwFWJy0QoWJDThJfEXGBkaJicoKSo1Njc4OTpDREVGR0hJSlNUVVZXWFlaY2RlZmdoaWpzdHV2d3h5eoKDhIWGh4iJipKTlJWWl5iZmqKjpKWmp6ipqrKztLW2t7i5usLDxMXGx8jJytLT1NXW19jZ2uLj5OXm5+jp6vLz9PX29/j5+v/aAAwDAQACEQMRAD8A+f6KKKACiiigAooooAKKKKACiiigAooooAKKKKACiiigAooooAKKKKACiiigAooooAKKKKACiiigAooooAKKKB1oAKKXqaMUAJilopcUDsHrR0x+dH1FLikVYO9OxjNJ68U4df8APNJlJBjqKUDkCjAz0wfrTgP89aRokAHHTHelAHPX+lHt24pwHXPUUi0hAMgnH40vGf8A9dLwAPX9aUAnHPv1xQUkGAevT+tKADgcnjn86UAZ/wAilABPr049vapbKSDHT/P6UuOCAKUAZPPTt1+lOAwcgetIpITAwCQOxHOPzpwwQev+euMUvoc5xz6frSgcDPv7fjUtlpCqATgHA4PHc+lOxntx09aAMEjA69enbPenYJwSR0z6duuKlstIdgEjnkDg/oelSDJ6D1P/ANb6U0AceuCOSelSKMD8SMfjg1DZokKOMnvjGalUckk5NMBHY8dcfzqQA846HH+RWbLSHqOQfb9enSp0wSOOeaiXGeevT0x/9ap1AJAA54rKTNUieNRkZxx0/wAKsoMEZzn/AD0qBAOMnt/XJ5qygBx/X61MS0iVMDPcevTipVJBGSAOvtTFGMc5HpTwD29h74z3FUUSrjjqfpxkdKlQnjI/DpUK8Y4P4ntUqc5/Prn8BTAkABOAMdB+v86lXIyfp9aiXPP5d8+tSoRxk9/8/jSGToDkevPf/PFXrcc8j0/OqKcEc56f5NX7cdPzqo6smTsjRhAwDjryMenSrkeAAMc9KqRcAA9uB/hVxBg5/r/KuuGiOKoyVfXvxTwRwcev+eaYpAzx/Sn4/Dit47HNIX05600k4+uaUkDtmkJxj2FWZjG6HjqOP8KibA9c/nUhJHJqI8cZH/1qoRE5JweO49aiYZyfqfX8qmY4H4/TioWwSPX+nbimgIHHTscdKjcAgnFTEcnPX1/pUTAknsOvP9aGxoruMZ56/kPeq0gAyTxx9O3pVtx1/Een6VWkAI46cj/69IZTkGBk5I6D1qnKO+D3z/n1q5KByDj17GqkgIJwD3I71DKRVYAk9u3Tv1603HTHT64qV1yevOffP40mDz9ef/rVmMjHGRjn3pvPI6nk5/SpMYJ49KaRyeP89s00iWxhwQfXp+NRNn0xgVMxAwe+KiY5JI4qkjOTuQMOoI7HpxULYzjHHr+FTSEjOffv3qu5xnj09uPeqRmQyEAnI/Xv64qtIxAPPOM/0qWVyAf881Ukbnp19efxpvQTRE5zjBxnuOf59qiY5zz7elK3fj8KYSDnPuRSuJoAeDye/wDkU7nB4PrTc+nXpn/9fenDg+pwR+OaBWHjHH4/h7c04c5PrTOgx9Rzn+dPUD+n44oESKcnjk/TOPqakBBx+AqMA84Gc849KkUkAZHXjoDimgJVB9O+OOO1PB/x/pUYOM59hj3qUcZBHUY/zimA8ZyOM556f54qVQMnj2/D1zUaevPAJPr6VIuOOM9aaE0Spg544qdegH/1/pUCnGfTPr+tTIAOvfHXtxWhLJ0BJA9BViPBx+QqBBz0qygAPI7fWmu4mieMA9x2FWIhnHB6fXnoKgjBBBPJ4q3HgYH/AOse9WmS1csRjOAO2Cf5GrkQBwfYH3qtF2PX/wDVzVyIYxjtx707ktFqMHIPXpVyNenPYe1VohwOMfTirsI4HH1oEtSzGoGOM9P/AK9XIh0461XjHIFXIx049qBk8anj/OasqOntUKDgVYQcg1IEiDpUqjpTVA4qRRnFSA8DFSAc0xakA/KgBQKXFAHX2pQOlIYUHv6Uo/lQetACUhoI6cUYoGNJ6UlKaQ96QIb1oxzSkZxSgUNmkUAHPNOAxQByKeBxUNm0UIATTgKUCngVJokIop4FAFOA9qBgB0pwoxTsUWBsB2pwpMdPzpfSgVxRSgUmKd6etOwmwo/lRRmiwhaKSloAKUUlFDAXNA6UZ5oFIQUUUenpQAUppKKAF9KKTP8AjS0CCijOaKACiiigBR2pMUUUgD60tFHagAoIoooAKKKKACiijFABijFHejvQAUY5o69qXvQIKMUlLQAUY5oNFABRRRQAUUUUAFFFFIAooo/nQAZozQKOeKACiiimAUUUUAFFFFAB36UvFFFAgooooAKMUUUAGOaOuKKKACj8KKKACjFFFABiiiigAxRjpRRQAlFLRQMPWiiigQlFLRQMSilxRii4CUUtJii4BRRRTuAUUYooAKKKKLgHHFGKKKLgFJilop3ASilxz7UYoATFGKXFH+OKBiYoxR3oxQAEDmijFFAgxRRRQMMUUUUAGKMUUUAJ1pcUUUAGKKKKBBiiiigYlLjFFHNABiiiigAxQO9FFABRjNFFABRRRQAUUUUAGaT+dLRQAYpKPrQaAD+tFA/+vS+lACUUUY6UAFBA5o4ooAKMUY/wo/xo3EH1pKWkpgFFHWigAooHSigAoo//AFUfhQAdDSf1pR+tFAAetJRS0AJSikooAKKKKACiiigQUUUUAFFFFFwCiij8KLgFFKKMUAJRRRQAUUfhRQAYoxRRRcAxR6UUUXAPWiiimAnr7UUpox0oAKKKKADrRRRigAP5UYzRxRjFK4Cf40YpfSii4CUYo9KX0ouAnpRjpS8UUwE9aKOAaMewoAMdaMe9LSYoAO4oz0ooxQAUdKOcH0o9PSgYUuPakooAMUfjmj09aP50AHtR260UUAJRS4o5oASlNHSjvQAmKKXH40lAgoxRRQAUUd6KACkx1paKdwEx0oNL2oxSCwnr6GilpKBBRilpBQAYooop3EwxRilxRii4CUGil4pAIKPagf8A16WgAoxR6UUDsFGKKWgLCUYoxRjmgBRSZ/OloxQAfWiiigAxRiiigAoNFFABRRRQO4UYoooAMUUUUAFFFFABRRx/SjFAB/8AqooooASlxQKMUAHekpf6Uc5oASl/wo//AF0Y6c0AJRjrS8UlABR2NLzSUAFH60fnRigAooooAO1FFFABRj9aKKACiijH50AIaKXtRzQAhopcUmPzoAKKKKADijFBo9KAYnpRS0GhCEope+KDTASiiigAo60UUAHFHeijH60CDr2ox1oo7igBKKX0o/WgBKKXFGKAEoopcUAJRSijmgBO/eilxRigAox0pKU0AB60hGKXFFAIOtFHWj070DuJS/jRRigAH60Gj+tFABRRQOlAAelFFB6UAGKB0ooFABRRRmgAx/jR0oH69aKADFHrSdRS/rQAdMUfj1ooHSgA/wD1UYoPbijNABRRRigAoxRxR3oAD1oNFFACUHjFHrRQAUfhR6UdxQFgNJ+HtS0YoEJS45pP50UAFFGKOaYBRQf896MH8KACiiigAHWjHPaiigA5pO9GaKAD6UHNFB4oAKKDzj0o60AFB/SjpSCgA/lS4ooxTAKKKMUAJRSnvSZ6UAH/AOqjFFFAAaPSj09qTFAB3ooGaKACgnrR+tBoAKPx4oNJQAUppOvbilp2ABRmj/CkosApGaMUhP1zRQAUevFFFABRRR1PSgA9KKKOKACkNBx+VHHSgA9KKPSj1oAOM0eneiigAoNGOlGKACiijPSgAz1oooxQAHH50GjpSd6AFo/SgUnp70AHrRRjkUUDsHHHvR+FB7UYoEJj/CilxSd/SgAFFHWigANBooo2ASilo60wEoo5/WigA/zxSUvvRjrQAlGM0vpRjpQAnFJ2px60mKAEo5ox3ooAP60hH6UpoNACUEdaXFBpisJjNGKMf4UEUBYQ0uKO1H4UDEoo60Y/EUCExyaMUuB+VBoASil/Ck45oAMUYoPYUGmAlIacf50Y/SgQ3H40UuOlBoAQ0EClHWkxQAYpMUooxjtQAlJ0p2OlJ+tMBMcilxRjp6UY4oATpj0o/lS+1HpQAhzikxS46UUAGOPwpM070pOtACYoP0pcUY5FAWE/Ck55+tL1pefpQAmOKMUUGgBBijFL6UYoASiigA0AJ3ox19KU/wAqTv8AmKEAev5UYox+dBoAMUnp60uOlFACdc0UtFADaMHFKR09KKAsIe/PSj8KXFJjOKBWA9BxSe1O9KTBoCwmOvtRilx+FHpTCwhowORR3FGBzRcBMUmMf/Xpx/SjHSgBopT3petJjp6mgBPQ49qTH507HNJ/KgLCUdMDFLRigBuMUUuKMH6ii4rCAdPxo4FL6UYoCwh70etKe9J1NO4WEPGaB+lL68UelAWEx15o7fnSnPA9qTGaLgAHXmig9f0pMZ9aAFpDml6mjmgBPWgjPSl6Yo9KAG//AK6CBxS46Ud6AExgGj145o+vpQRk/wCRRcA/pQaOveg0AJjpRSmkwPT2oAMdaT070p7e9HB7c0DsIR/nrRnpS45oI9qBCdc80UvrSfhQAmKMUvTNGM5oAQ0HrSgCk5/GgBKPTj3pR2/Og8np70AJ/PrSfhThz+lJigBP5UGj0owP88U7gJ3pDwBzTiPbrTT349qAGMBgcf0qM9+PapDzmmHgjjimgIWAOeB6VEw54/yKmfr+NRN9elUgKzgkmoXxg/l3qdxgnv8ArVdwO5Pr6VaEVpCOuD7fyqrLjnn168Grcox7dT6VUl5BGPcew7/hWiJZRlxknB9fxz6elVXGMnGOg9atS9M/y6elVXPBIJOfw+orREMrsMk5wT0/CoiMEnoOv+NSv3yOpH5+lRkHJOfX2/Kghje4yfUf5NSJjPTJ6+n40zAJIxj/AD3qWMYxxTQFiIDB7Y//AFdqtDp1/lVaMnjjpxz6VaXp1NIo+eqKKK8I3CiiigAooooAKKKKACiiigAooooAKKKKACiiigAooooAKKKKACiiigAooooAKKKKACiiigAooooAKKKX8KAEo/ClxRQAUtAOKXg0ikg9+9L/AProHpS8gCkykhR/ntSgDFIBinDvQaJCgf596Udf8k0DgdPalA/DP6CpLSAAkj/PengdM/SkwQSPyHtSge/Q0my0hQOR0/yaMYIzzjj0pQMj8f0/xpwGMnH50rlJAAeMdenrx3NKABn2z/kU4DHcc5/PFKOuPTjHI/LNS2UkIMHtwPp1680qrjP+RRgnoe9PwckZwOtJspIRQCTx9M/lTu446fX88UADr0GM5pwJxkDgCk2WkLg46c5PT+tPCDjuffnmkAJ5zTxkZ5GRmob7FpCqATnngYFSY4OTnvzzTQDngcZHanjrwfX09e9Q2WkCgnv7f/r+tSqAD0PQD6n3FIB0Htz0HNSKPmz1Oefp2qGzRIeoBA9fX/69TJwR6Dr6YzUagYJx0/PGamTkgfl7VlJmiLEYzgY64/Gp1HIHt3qKMdOfx/rU4AAH596EWiRcEjA+lSLyM9P1qMYGPT39fepV9MelMpEi88EDPJyKfnBJx79Md6YvTkY5xUgPJ7dBj1+tMLEi4OOBn9c1KOSP/wBX9ahB6evriplHByMc0mBPGPcnP4mtG2ByOPx5HHvVCPJI54HHp9K0YASBn1A9KuG5EzQhA49ePfBq0o65P5VViBOOcdPxq0vbHH9K647HFMlXqB7Z9acD0559P5UwZzwc04H3569K2RzyFJBBppOMdOmPSg9u/NNLEjn6Va8zNoaSDnrx6fWo2AyMkjrTyeck+oGP1qNjnJ/Dtj8qdwsMbHIAJ96YxPJ+vP4VIcEEc+hphB5x7jn+dNMLERBOR+f1qJwCTkdOPw9qnIyevGajYYII7/54pgVZB1//AFcVVlBGfTkfhVuUEgnHv1qrICeh6ZyanqCKcgOcEdx3/WqrrnOQO4/WrkgzkepOarSA8c8f1qGzRIrlckj6HpTSgHPofX371MUBxwcnOD14xSFQCRwfrUCbuQ4HGBxz2phHJyeM/X9anK8Yx1//AFdqYyEAnrk49x+FUiWVWAGQcdeO+PrUT9//ANdWXBGOvr/n3qBwRn8zycH/AOvVJGbZVkzkjt/ntVWUnnJ9CecVbkAyeM5/zzVOYZBz7imQVZjkHjj/AD3qm5BzyOn61ZkPJ5OPc/55qqxJJyPX2ob0AjJB4Bzzj05pvAxxjB/z+NOOCM5x2696aQeAfc/rxSE0JkHAAI5/D/8AVThwRzx7/T+dIMknnHP0/T2p3Prn29vWgVhRjHc9KevAGPr06U0ZPQ+px/8Arp444ye3FUKw9TyTn0z2+lSAk8Z75x/jUa4yQAQe/oeKlHQ5OPahCHqDgd+p+vtUq9Sf8/SolyD0/LHTsakHHocdsnNMCVBnB+uPUfWpVzzzz2qJcHBxnkY5/HmpVI6k+nHT8qaAmXgdf61MgOSeuMf5xUCnBHf2zVhSSepxxjtV3JJ07Ad+PXvVhO3rngk5qBewPbmrCDkDp1OaaBosxjHX8f8A69Wo85HA7E+lV4wRgkE5OatRZAHp39x71a8iHoWogDj0HH/68VdjXnIHWqsSgdvc/wAzV6JQSAPY+tUieli1EvQY46+tX4RjAI59cVVhGCM8E/iKvRDABHHShhaxPEmcfl3P+RVyNcYH0/xqCIdD0q0gBA7/ANaTYiZBwOKnUYxUSD261Oozj8KkCRRUqimKOnNSL/OkA4D86eOB70gA4+tO7UmAvp+VO9aSlFA0JSmig/rQA38KQmlNJSGNoxS4NGKGy4oMdKcB+lAA9KfiobN4oQD/AD05p4FAHtTgKlstKwY/wpwHSgD604DpQUAFOA4AoxSgUhABTgKTHSnCmDAUooFGaYhaXpSUZoExaKKM0AHFFGaKADvS0lFAC0Un+NLmgAzR2ooosAtAozQOlKwgooooAWj+VJRQIWijPWigAoope1ACe1L2pKWkwD8KKKM0AFFH/wCqigAooooAKKKKBC0Cko6UALRRmigAooooAKKKKACiiigAooooAKPaiiluAUUUUwCiiigBcUnpRS0CCijvRRYAooooAKKXFJQAUUUUAFFFFABRR6UCgAooooAXFH40ZoxQAdaT0opcUABopPWgUALSUuKMUAJ6UuKO1AoAPxpKWjFACUUuKOKAEox1pcUlABRxRRRcAxSf5xS0U7gJ2opcUUDEFFH86P8APNABRRig0AFIe9LRincAoxiiigA60Y/GiigBCPajHSlooAQ4oPalNFABijnrRRigBMUUuKTHNABRilx+f6UYoC4lFL/+ujFACUYpcUlAXDFGKMUvSgYlFH+fSigQYooxS0BcTFGKMUUDCiiigAooooAKKKKADtSUtFABikpe1J6igAo70UUAFGKKKBCc0UvTtRimAgooooAKKOO9GOtAAKKKKAA9KDRiigApO3pS80UAJ+FLjpRRnpQAYoxRRigAx1pKUdKMUAGP8KOKOKMe3WgQlLij/wDVRQAcfrSUuKMe9AB+FGKKAOtAB1o6DpR1ox1oAOKT0paBQAYpKU0fyoAKMUlLigAxSUvak/rQAUUpoNACUUv9aKAEopfT0oH6UAJjNFLikoAKKU9aO/WgBKKUUYoAQ0e9L2pKADFHeiii4BQcUUUXGFIaX9KKLgJRj/Ipce9Hei4CYooxwaWi4CCjvR680fU0wCijFGaACg0YooAOtBox1ooAMUUUUCDFGOtFFACUYpcfWigAxSYpRRQAnWj0paSgAoxS4/GjFAWEpDTsUmOtAgooooATFHb9aWigLCf59aXiiigLBR0oooAKKMUtDAT8KKWigAxR0xRRQAYoxyKKKADOaKMe9FABjpRRRQAUUUUAFFFFAwo9fSiigAoNFFABRjFFFAIKKKKACijFFAAaMUUUAGOlHNGKB2oAMUf/AK6M0CgA5pOlLjiigAoo9fypKAFFJjmloNABik70v4e1GPwoASilxSUAB7UUuPek70ABHWiig9qACjHSj9RRQADFH/6qKKADFA7UUdqAYYpKKXFAMTFHFH+TRQIMGiijvQAlFLR60AGKSl75op3ASilFHtRcTEFH8qMUUABoHaiigAxR+FBGaO/0oAD0o780UUAFFGKMUAFFFGKADFFFGPzoAMUdKMdKKADn6UdKMcUUDDFFFFAAO1GOlGOvtRQAUUUY/Oi4BRS4Pr70YouAg6UUoBOOKcEJ7UrhcZjpRUgQnHFL5R9KXMguRUYqXyj6UhiPpRzILkfPNJn8s1JsIzxTSpHammguJijNGDRRcBKKX8aPxpgB6CiigdaADiijtRmgAxQaMUGgAPWj9KD/ADpDQAUUUUAFFFJmgQtJ/wDqoJ60fyoAKOho9aKAA0UUUAFFBopgGKP8aKKADFJjrS0UAJiiijH8qACjtRg8UUAGKO1BooAKQ0UUwA0UdKXFACUD8qKOaADPtR/Sjig/57UAH/6qQUtJQACijHejNABzR6+tJ+NLzRYA/wD10UZozTAOx+lJRniigVwoozR+lAXCijHWkzQFxaO/0pM0ZoGLR60en50np/8AqoAMUUtJ/hQAZopaT2oAMUYo9aKAD1o5/SgUUAFFFHrQAUUUUAFGKMdKCP8APSgAoz1oooAKMUYooAMUn8jS496KAE70d6KKACg/SiigA9OKKKCP50AJjrRS4/xooASgj8hRj1pf/wBdACUevvR1o9OKAEopfWj/APXQAmKOlL9aTHrTAQ/1pcUY5ooAQ9zR/n2pf5UdKAG46cUelL/KigBMfjRS+g9aT/8AVQgAjpQRj+dGOnNFMBKKXHSj09aVwEopcUmKLgJ1pcUYo/pTFYQ0Y6UtFAxMUYpaTH6UCsJ+FGPWl54oxRcLCYoxRRimFhD/AFoI60tGKAsJjNBGc+tLgfnRigLDcUEdKXtiigBOaMdPyoxRmgQYpMZpx70mf1oGJjkUEcilxRQAmOnHWkxTsYzSUxCYz0ox+BpfWj+tACUEe/FAox70AB/lSe1OxSY4oASjHTjNLjpSdKADmiiigAPWj044ooNACUfoKXAoPWgYmOvpSGlxxQKBCYopccUmPWgAxRRijHWi4CHtRjrS4H1FFACY60hA6c07tSUDEoPftS9c9KQ0CDHtQRyaMdaPTFAAaTHXHWlOOKO9ACdOKMcUYoxQKwH9aQ8j9aXHT/8AXQKYWG45FB6076+4pPwoAT9ev5UEDoaXrSCi4BikI96XrRQAmOTn60mPenAcGkoCwmKXFLzSYoATH6UGloHP0oCwmOlBpaSgQmOaDn86XH60dKLjE469aB/9ej0ooCwcZpMdfxpT19qO36UAHUnp/KkxS0cc0wG45z6UYpxpD16/jRcBCOuaBzjml9aQ/wAqADFBHT86KAM5NACY/L+tGOlLgGjvQFxO57GkOeaWjtQISg9D70tIKAADr9aKD09aMCgBOKPX6YpT2/8A1UnfqPWgAwefSkpaOv8AOgBCfxoP6flS9fpSUAJjmmnNOH+fpSYPpTQDCBUbDrUh/SmMOvemgZE1RNnke2albmoX9vWqQiBxz069/wDPaoH6Hj3qwwIyPTPT9KruDkkf5NWgZWkHJHXn/IqpIMA8H+h/GrjnJ68cmoJcBSe5yatEt2M2UHnAP5Z4qm5AJx7YxV2fkk56k8dvQcVTcAE9Oa0Rmys4AJ5qPucj1z9fbNTN1PPqPSomAGT0J9fy7UyRoODyPWplOSPyqLn09xUi4znHcf5NO4IsxYBGDnP4j9asjp0/Sq0QyRxgA4HOOPr6VZGMcmpY0fPdFFFeGdAUUUUAFFFFABRRRQAUUUUAFFFFABRRRQAUUUUAFFFFABRRRQAUUUUAFFFFABRRRQAUUUUAFA60UdaAAUfhS5oFAwFLikA/WlFIEAAyKcDSA54pRQWl2FHp2pcdM/nR6f5zTsZpNlpABk0oGePxz/8AWowSQOaco5z6/jSLSDAx+lKBkUYzgY56f/XpwB54+lJstIMAc5/xp+M455Jxjn19aQgD8eKUA8kdufrU3LFHBII4zwf0pVAHBP58jFGAcc9BTgBzk+mDSbGkGBknHXB/HH8qcBkjj8M0Y655z/kU4AY/+t/OpbLSEwR3zj9fencjOPTGenPXjNLjkgkdf0oHQYA+nSpuUkOGcEkjnH+eKUAnr0yeO2PahQccjHrT15HTk/zpNlJAo445605Rg5xyc/40g75HbH456ingH1HOPX/OalstDlAIH45/OnAD16j9etJkYHfrn6+tSKBknHrz+HpWbdjRIVQCR+JqQAnt34piZGcY5wO4xUozjI5B/wA8ZqGy0SKAD3wcZ6f07VMgBwe/eolBwCMeuDVhAOMDv0rNmiJ0GAD+GB2qZe/bp/kVEo79z04qVSAB7dulUUSKOT78mnggHA4HuBUa8YOc8/WpFz/nqaYyReOc+/H6/jUg6nnnHWo1OD0+h/z3qQAn1zwP8ikA9cjHP9f5VOmTgkcDnOcfhUCknAwP5VOhPAx35oKLUWcgAcEjPvWlbjgen+etZ0PXuent71pQDgHp0961gjKbL0ZGQOuMD9KsKcnkcfnVZMDH4fjU6kg8jrg/rzXVE45koJHfP4Uuc9f8/SowTk+lOB4HHP6Z9a1RjJDmIOefamH68df1oJODzz1z17dTTc5+nFUmQ0BJ9cdqbjk8+lLnJ9Dz17UhIx+P+c0xDfXn0ppXJI7cU/IP0Pr+fSmnv6dKdyRjDHHbgeveonGCQPwqVsZHTr+GaibAPSi9h2KzjOefXJ/Cqzjrxn6fpVtsAHH161Xk7nPuf8+tS3caRTkHGM8Y/wD11XYEk8dDmrcgOT+NQsmSOee+fzxUtlEITAORz+dNKDp64JqxsHB64/nTWTAGB6HPX2pJCZXZOvqMcdPxqNlyDnoOevP61Z2DIyPSonXk8kceneqRDZTdSCTzj64/Sq8gznnp6DFXZE4znJHb/CqsgySMfh1qkiHqUZQBn6dT61TlJ5wCep9fzq9MvJ6j3qjKDk4z09BTJ8ihJ3565HXGDVduQDjHX8/arbggkE9eKrsoz0/z/hUsCI4PHpxj8PamEEEc/icYFPI5PT3Hf8KTGQOOO/XH/wCukIbjn/P1pxABBI6f5BNHUD1yMU7B5J6/pTuJoBnjj8f8KcpBJycfypuBkHBOee/SngZ6jqR/k0xDwAeQenpUinrz6H1pg6Afj6GngjI4x/8AqyKAaHLxg9T0H0+lSjk+561GATjnrz/+qnjtngevWqFYlQYxx1Pf6cVMnJGD1xz1747VEMYHPXPv7YqVQcn+g/LmmtBEqcEdxn/9easKBjpjHSq65OOD1/GrSAZAA9f85qhWJ0HQ9uO38qtRg8D0AHp+NV4x0/Pr+HSrKDB5PbGaaBlmMEgAn0+v/wCurkQGRjPt35qpEACcdfzq9EBkEcYx6/pVolotQjJAz+PtV6EEH346+tVYVJ7A8jt+P51oQg8fz71dyGi1EvTPX/OKuxA4FVYgTj6DpV2IdB3oYixGOn+RVpBnFQIDxx/9arMY4BqQJUFWFA44qJBgjFTKDSYiRBwDT1pqjp/k1IBnHFIBwH9Kdj86QAfWlHX0pMY4UUAdPSjpQAUGig0DGmkx+Jp3rQB09aRSQgGacBigDpTgOlS2bRiIB04pwFAB4p+KhmqQgHSnAUAU4CkUAHSnYxQBSgUCAZpRRjNLTAB+ZpfSilFMApaSigQtHakpc0AFH9aQUuaADNGeKKKBBS0lGelAC0UUZoAM0tJRmgBaAcUnpS0CF70ZpKKAFooopWAM0ueKSigBaKKM0CClzSUv40WAO1J/KilzSsAUUUUwCiijNIAooooAKKKKBWDNLSUuaAAUdqOKKACiiigAooooAKKKKACiiigAooooAKKKKACiiigBaKKKBAP0ooooAKKKKACiiigAoFFAoAPSj+dFLQAd6Sg0uaADFFFHSgAooooAKKKKADFGKKKACiiigQUUUUDuFFFFAXCjFFFAXCk55paKAExzRilo60BcSilxRRcBKKXFJQAUYzRRTuAmKMUtFAxKMYpaMUAIaKWii4CUUuM0mKLgFFGKMfnTuAUUUUrgFGKKKYBiiiigAo60UUAFAoooAOKKKKADFGKKKAD2ooz0ooAP8mjH+cUUUAGKOgoooATijvRx+FHagAoo6UUDCjtRRQAUYoooAO1FIaX0oASijFLQAlFFFAgoxRRTGBpBilNFIQYpD/OlopgJR0paMdKAEopaMUAJRS4pDQAUHpRRigAoxRiigAooooAKKKKADrmiijFABRRiigAopcdfSk70AFFFFABRRRQAUUUY6UCCjFFFABRRjNGKADrRR6UUAHFAoxij0oAKMUUY6UAGPyoPSiigYUUYo7igA9+9BooxQAYoxQOlGKA3A9aD+tGOtJz+VACmkpelB60ABHpSUUuKAEopcflQaAEooooAKSlooAT/AD+FHpS0UXATHvS0Ufh7UwEoNL+NGKAEopfwpP50AFAo7UvFAhKPTij8P6UYoAKKKOhoAKMUUUAGKKKMUAGKO1FFAWDNAoooCwY96SlooEH+FBGM0d/xooAPT86KKXPvQAg/xooooAKMUUUAFFFFABQKKKACiiigAoNFFABjPpRRijFABRRRmgAooooAKMUUY6UDQUUUf4UAFFFFABRQccUUAGOtBoooADR29aKKACg/lRR/OgAooo/HigAz1o/IUfzoNABR/SjmigA9KMUlKaAE70Upo/lQAlFLSUAFGMUf40UAFFA70UAFFFFAB2o7/wCNGOlB/WgQlLij0ooASlHakx70dfrQAUUD3o/pQAUY6Uvak60AGKP/ANVFFABig0UUAHFJ6+9KaKdxCUYpcUcUXASijFL6UAJRS/zpKAClo/nSUbgHrRRijpQAUY6UtJjigAooooGFL69KSlxSAT/P1pcUoBOMCpFjJxxSbSC5EAT+dOCE9qsLEOOKkCAY6VLl2JcisIiccVIsPtU+AKWpuxOREIgOtP2Ads06imK4gUDtRgelLiilYLhgf4UmAe1Lijt0piuN2g00xKe1SYo6UDuys0HXFRNERnirxpCoPWi7Q1IzyCO1Nx1q60QOcCq7RYzxVKXRlJ3IfWlpSpH4UlXuMM9PaiiigA+tGcUUgoAXPSjrRSZoADRRRigANFFBoADRRRTsIKKKKACiij09aACjNFGaACijigUAFHvRRQAlFHeigAo7UUUAFFFGKADOaKSjNMAooooADQaKM/59KADPWj/9dFGc560AJ3/WiiigAzRRR6/lQAccUhoop2FcP/1UUp60lABR6UnSj+tAC4oP8qQ0tACUZo70UAGaBRRmgAo64oooAKKKBQFwo96BRQFwJ60UUfyoC4UUCj+dAwzxRRRQACiiigAo44o7+9FABRRRQAUYo/nRQAfpR/OiigANJS0Y5oAD09aTFLR0+tACc/hRilxR1xQAlGKMUdzQAd6OKKKAE/THNFH8qXFACf8A66KXFA7UAJxQaKKAEwKDzjtxSnA7Ud6AEox+VFHU0x2Ex9aXFFGKBAaPSgd/Wk69qB2CilxSUCExRS0UAJxQB1paSi4BRRiii4BikI/Cl/GjHNFwEopce9FMBpo6/wAqWjHSgBOtFHr7UY46UAFGKMUUXATHWgjpS4o7UXATFGKX3pDTuAEfhQR+FFBoFYTHajGaXFJQFhOaKXFFAWEoNGPSj078UAFFHrR3H5UAIaQU4jFJjpQAmM5pfSig/wD16YB6Uh7f/rpfwzRQAlH86KX/APXQAmKT1pcfrRj+tACUetGKKADpQR3oooASl60Ud/8AIoAT+VAzS9KOaAG0EZxSjtQe9FwEo54o9aKLgB7Uh7Uo/wA9qKLiEIPNHU0pox0oAaSP8+tGBzSkdaD2oASjH4Cl9aPSgBMUlOxSdz7UAJjrQc0ppKAsJjr9aAM0vvQe/wBKAEpMCnHpQOSKAG4Bopfyoxz9aAENFBxzxxQR0/z+NMVhMc0e3+cUuKMUXATHWkxjNKRnFGOlACH+VKe9Gc4xR/8AroATjijpR/PpQaAD/wDXSHtxS/pRj8aAE4OP5UGl5PPSkx0oACBxSHr60tGCKAuJntR69OaU0df50XAaaPXnFKQaPbvTEIfx7+9GBzQepooATHSjHB54HFL68f40UAIeKO/vQeo60dKAEGefpQR6Uo4oJ6cUBcTHSilxScUAIf8A69BpfX86Tv8A56UABzxjjNJg/TFL17c0cfnQAn4cU3H6cU8gU30pgMYc8VG3+NPIx2phzTQETetRNknGOP0qVs5PPtUTnA9qoCFxjPPtVdxjJqw3Heq7n34/KrQmVnA9c5qrKeSO44/WrUuTnA//AFVUlBOeOB/hVolopTY55GTn+dVHyM9c49sVelXP4/jVNxgnrgdPpWiMysSMdOef/rVCR83Izg9P/rVM3fnpyOvWoiMnOMeh9qZLEJznjHP+etSR8n8u3Wo88D398c/U1JHnnnng/SmBZTPB6nr/AI9KsqOOn6VXjBwMHpz/APWqyoG3/wCtUso+eaKKK8M3CiiigAooooAKKKKACiiigAooooAKKKKACiiigAooooAKKKKACiiigAooooAKKKKACjFFFABRS0goAKWiigYuBnvRjBopfakNIO1OHNJjPNOFJlpC4OelLgn27Ug4NOAJ+mTSNEg7jueeelPA4HP+fak9Bjr1709cHHQdQMiky0Cjpx+tOHJGCc8D8aRQcjnA9/WnAEAY+vp2qWy0LgEkY5+nNKARjP8An0NKAACc8joKdgEDI4Of85qWykuo1QD0HfHTtTwMEHJ4/LNAAGQP09fSnADjIyAB70rlITjJ6ccZ6frTlAyc8dPzpQOeOnbnP4UpByeOnHPTFTcpIFAJ65x6H9KUDg5JIABoxjJ47Egf4U7A49eM9uKTZSFGD0ORzTsDgZGfrj+VJjGM5zx3OfTtThgYweDyPXr6VLZYoHJ7jp64/GnAdgRx+VIM5Bz1z6H6U4ZPGSeo/H0qWykOGAQPTH48Y7U/AJHX2HXmkUA5OPw/GngD16d//wBVQ2aJDlBHqcdScGpF75Hv26+9MAwBz68g1KuAQDz+vFQ2UiSMc4xxVhACBkE/59ahQEHjkZx/9bFWFAJxjHGf8+9ZpamiJU9PQ+uTn/GpBnAOPXk/56VGAMYz059P8mpFAwDj+fSrKHrn1yP8ipADn2A7H8qjBxzyTyOvb2qQZznp2/8ArU7DuSDOQM4I/wA96epIHXp64qMEAdcnjPf9fWpOACfUgc80WAkUEEZxjGPoPwqdBkgZ4J7ZqEAZPPP6VNETkZz2x9KXUouw9R35/rmtGEZAx35PvVCEdh3yeavxcjJH4D6VtAxm9C4nB4PA59f1qXPTB5+uc1AhIxxUg45/P/8AVXQjlkiTOMfTGPf3pxIIGOc5qMEYPOe1KDj0zx1rRMyaFycgj+eKUkYz3ppx6cdcdaMgj06VSZDQ7gdz60hJOevtSZAPXOOKXIBBHsDTuS0JjAPPfNISOePQ0uRjtnPXp+tNOBnB9qYrDT1GenXt1qFgc5z3xUp64PuevFRMMg4/DueuKTY0iB8E9ffpVd+cge1WWBOM/WonwcceuP6VNykupVYZHT25H60wIcnA55OT6VOQDnken4U3aDggfl69KlvULEW3Axn17fzppQ4HGBVgLkkYyc00oCQBnPJqkSVmHBwMD04H4k1C6jnjI7VcYDJyOCOe1QyAZAAwOvY1Rm3qUnUYORgjHTkVUkUn68kZq+4wDjr061UkHJ7+nvVXFa5nSrkkkH8On5VTdASeDx+BrSlTg446/wD6qpugJP5+tNIVjNkQZJPI5HvVdkIPXntWhLF1OPf0qsyDkEZ60mg0KZU5xj/PWmlcHr79vSrDR9SB357Y4pgXnOO30qQsQ7cHB6HPvQBnIx05/GpNuCPX86NuCf6UEtCAZz7596AMgkHP689aeqgZPbp/n2oCjGAR+HP86YmhVAIJ78U7HB68evYfjSBckYOCacAcgj/P0piaHKAeh7Y+g+tSAgdyc49P5U1c46dMU8DGOPXp1pisSA4x6fn+dSoQSDnk8d6iXBIGOuKmRR0xxxVBYnU5xge/4/jViMEkEjgZ/Cq6DgHsR9O9WY8kg5wBx/hTTFYsxg8HB6j+dWYxwOQep9eKrIOcZ9RVuMYx+XpVITLMagnrzxz04q9EvTjPYf41VjAIGPXHFXolyQCO9aJEstwKTge4Jq/CvHrgfTvVWFRkGtGJRgcZ6YqiWWIlAx+WParkYH+fWq8I68fSrcY6Z9BSbIJ0BPT6VZQYx9PpUKDpx6VZQcjjmpESIOlTKMVGg71Oox9aQ0OA/Onr9Kao9qeBmgQ4DNO6fQUgp2PakMMUY60cfhQRmgAPekpaMUhpXEAzSgY+lKB0pwFS2bRQmKcBRjp60uKk1SDFOxRinYqS0hMH8adjvRjpSgdKYABSigUoFCAKXH+FHej0pgFL680npRTELRmko/nQAv40ZpKKAFNApKKAFpc02l9KAAUtIKKAFzRSCj1pBYdmjnikzRQKwtFJmlFAC96KQUe1AC0Ud6KAF6YopKKBWFopKKVgHCjNJS5oEAoo4ooAKKKBQAvaikHaj0oAWiko9KAFoozSClYBaKM0UAFFFLQIBRRmigAooozQAUUUUAFFFFABRRRQAUUUUAFFFFAC+tJx1oooAKKKKAFoo46UZoEFFFFABzRRRQAUGijigA/zxS5pKMUALQKKKACgdKKKACiiigQUUUDNABRRRigAo6UUpoATGaOlLigUAJ9aO/NLjpRjrQAlGOKU0fyoATFGKXFFACUUtBoASjFLikxQO4Yoo/woxQFxKPSloouAlFLikxTuAUUvSkoAKKKKACjFFFABikxS0UAJ0oxS0UAJ3oxS0UXGIaKMUelFwDFFFFMAooFHWgAooxzRQAUUUUAFFFFABig0UUAFHvRRQAlLR+OKM0AJRRiigAooooGFFFFABjrRRRQAUhxxS0UAJRS0evrQAmO1A4pcUUAJRSntRzQISilxR+NACUUtJ/KmAdaMUe2KXFACUY6UvNJjpSAMUYooNMAxxRij9aKACiiigAx29KMUdPpRQAYoFFGfegAo+veiigAoxRRQAY5oxS4pKACjFFGOlABijFFFABRjpRS0AJijvRRSAKKKKYAQKSlx0ooASlxmg0fyoASj+VL/AIUfzoASilxR9KAEopRRigBKKMYox0oAMUHpRjtRjmgAo6UYo/GgAxRRQOlABijiig0AGOKSl/nQaAAdaSlxRjpQAlFLj8qO4+tACUUUo7UAJRS46UlABRij+lFFwCiijHX2oATFLijijHSmAc0cUUUriCkx9cUtFMApMUtFACf40d6WigLCUUtGOlACd6MUuKMe9ACYxRS+lFAWENGOtGKXr2oEJjij05pcUcUAJRR60YFAB/jRR/KjnigApeaT+dL6cUAJijFLxRzQAUlLSUAFFHp+VGOtABjpQKM0UAFFLzSf1oAKKKKBhRRj3ooAKKKPSgA9eKKKKACjpRRjpQAUUUUAGKBRRQAZooo/nQAf1ooooAOn40f40Y60AdfrQAf40c/lRRQAfyo/zxR0oxQAUlL1pKADA/KijB4o70AFFFFABRRijj60Awopce9JTuIKMf596KKQBRRRQAfhRRR1xQAcUYoxRigA/GkpaMUAJRjvRj/ClxQAlFL60elAWEooo/nQIDRiiigAxR6UUY6U7gHrRRjpRSGHpRiigdqAsGOlHWiigA604KSRQBkipkTGMjNJuwmwRBxUyqBzQo/M04dqz3JbFx2o60daDTJDtRS0UAIaWiigBP8APNFHtS0AHakpaTgUAFL2pKWgBPSjFHSj/PWgANNKg06ilYaZWeLr71XZCDWgQCKieMHJxTUmikylgjFJ/wDqqVkIJ4qMjFaJ3K3EpaTH+FHp+VMA9aDwOtGaKAD+lFKO1JQAUUGimAUCijNAgooo4oAKKOuaDQAUnrS/rmkoAXNJRS0AJnmjHTmiigAziiiigAooooAKOKPUUZpgJ75pfxpPX35oNAB0paQ/1o47UAHSj/GjiigAxSd6Wk9KADGKKKDQgEoGfSlpKYmLn/GkoNFAAf8A69H+NBH4UUAHr2pM0tJ/OgA7UZzRnHaigAz0ooooAM5oFAooAKP0oo65oAKKKB2oAKOmKM4o68UAFFFFAw70UZ6Ufj70AHr0oooNAAM/jRRRQAUZxR9aUdqAEoFA4xRmgAoNGOKKACg0HPFFABRRRQAY5o7UYoNABRRRigBKKWj0PagBOtGKXH6UfzoASijFFABR2oooATt1pcUelGOv50AJjpRjrR+lBoAMUYox09aD+ZoAMUlLRimAlB70vpzR60AJikx0OcUvr70enFABijHSjHSigBKKXHWjHWgBKTFOx+tJQAhopSPzpKADHeg0Z9qMUwDFGB9c0UGgBKMUtHSgBAPwox+VLik6Y6ZoAMUUYo7UAJRmlHak57UAHtSHjFKaD3ouA3HJH40uKXvSY6U0AH9aMdDRRjmgBKKKCOnpQAnrRj86X0pO9ArB3ox0pT3pP50DDmjAxRjpR6UbisJjpRjpS46UGgLCY60UDtxRQFhD0oH4UtGKYxMYz70elLikwOKAA/yo+tFBoFYSilwOfSjFIYlGKWk9KYrB+tBoo9OKAsGKT0FL19qSgAx6mkIIpaPXigBDQKWk6k96ACg/zox+NHWgBMUfpS/5+lH86LgJ6UYooouAfjSHvRjk4+lFAgx1wP8AGjFLSHPFACUY4NLg0nf/ACKACjGc5xmjBoxQAhpCDTsY6UlACYo/pzS9xQR7UBYTHA70enejHvQR+NArCZz2owOD6UoHrQPxpgJ60mf0pT/9ag/4igBD+dGKUdqOaAE/z7UdKO3Io/8A10AJx/Silz+HekoEGOv50elBwDQentQAnXOKMYx/nFHTtRzketMAPekFL+lBFACY96OKOf8AP8qPX/8AVQAnBz9aP/rf/qpcUH8qAExjr/8AXo45PSjHFHFACHv+VBpT2pDzQAHnIo6fzox0o9aAEP8AKijH4GjPtQFwPemntzSnqfpSHH0pgMPeo2xg/UVIwGDUbY54poGRsM5PvUTAc/4VKeM1Ex+nU1SEQOCc96rvyD6CrLg4Pb/Gq7AD/OatAyrIBz1H+HWqrgDPXPP+cVckx1xmqkoI/XrVollORgARjuRVJ8jPPTnGKuSADJ+vbv61UkwD6VoiGV3ySTk454FQscgcEfr9R9anbBIB69TUR5JxnjA9OPWmiGMxx+II/nUkZBz65/WmDqR3yMmnoBkEdDjp+VNgWo88cHrj0qwOnT/P51Xj5AA/Qf54qwOnX+dSyj56ooorwzcKKKKACiiigAooooAKKKKACiiigAooooAKKKKACiiigAooooAKKKKACiiigAooooAB1oooNAAOtFGKX8aAD0pe4o44pc4pFJCY5FOAoHWl7GhlJCgdP880oFJjrmlAP9KRokOAyTmlxx7fWgdhj86cO3HvzUtlpAAOhxxThjBx+dAGATgc9f8A9VOAAxnjr19KlspIUA57gf59Kdg9R9cCgDBPB9OvSncjPtj/ADzUt3LSADg9z15pQDnnGRz6j3o4waXAIOP/ANfPNK5SHYPAxj3/AK0uME5x35oBGTnGaU4JOP8ADNSygwRjn19vzFOGRgevJ9KMA555GOnJzSjBHXn8qTZSQoAAyPQn1+mRSjBHU/TP6/SjHPA64/xNOBODnvgY/wAfpSbKAcgZ6/8A1+9OAHXI4yTSAEngZNOAOAepBz/jUstCgAjr+VOAA4z2wKRcZAPv0/xpw4JAz7fT3qWykPAwOuMYHHbmnrwAOo59jTMZz3PGR1p6kEgdQO9Qy0OwcjAzz16VMnGeuf8APSoxxgA+nqe/OKmQDjBPH6/WobLiSxjv6fnU47gdCD/+rNRID6+v5/4VMpHPOaS1LHqDgHPT2zUg6f8A6xTFIyfwPPFPHOR26nt+NUUPGScH6nn+tSdQOajGCQf/AK1P6fQHPtTAkAHAJ68DjP61IuMgdOh5/LpUY55B4IGf8PrUikkAHt07n1oYIkTGR+XP86sRc5Oc5/GoF5HTnrViLkggcYGD/hSWrGXrcDI5z1NX4+B15GaowjGOe/P8quoSAP8A63XFbRRlMsKSAOQT0NSAkc9TyKhU56exp4bBH169PxNbo5pIkyeM5x7/ANaUEkjjPf8ACo8k5yfegEjGD2PHPSrRFiUHOTn0HtRnn06D29KYCckHpS7hwM8c/wD16q5LQ/OAPxzSEjnPP+NMDDJz+dGSSfy/yadyGh+RkA/Sm5yTzjqKTJzz/wDXpCTgZPQ/T86LhYGyOM+p7Go2PvzwTzTyevP+e1Mboeeh5FJsLETVGw4BA6fy9KlY8ZOBjNREA5PJ9ulS2MjIznjHc96NhAHt/k0/AI6Dr/kfWngDB9sZpJiZEAOePc0hXGCc55xj6fzqYqME9Ov/ANamlQQevHPrTRJWK8EYwOvOAfeoZBnA61bYAE5H4etQSKcHj2/yfWruQ0UJR19O3+f5VWkUnJA4ANX3XOcc/wAvrVd068dev+FUmJozpEzzjr+H5VVkjyDj/D/IrTkQnrz2qs6Dkf5Pp0p3sHkZjx8Hj14qs8YzjH1zWo8Zyc1XkjI4A7HntinfuK3czGTqenXr3qMoeDz2z249MVedCPbtz/SoWjIJwDgevPNS0DKmzkcf0/yKNvJ4wPQGrGzOeMYFNKYJ45/yelICDaSQM/Xt+dKFIA9ean2cAgA9ulIU64Pt+HejUloi2kjJHoBj/PSnKCR06d++KkCHIH16f5707acY/wDrY/rTQrXIwDzzxyR9fSnAHg446Yp+wnHseaeqEdueP8kUxWEUHI4/mfwqdB1x7H3pqrjnAHSpVQAjHc4/GqBolQEY/l71PHzz0zk1HGvXPHIHP51YjXOMdOB6U0ImRQcZxVyIHjP0qvGuSPb2/X61diUggnkD26/j61aIZYiXBB6A8Y/rWhEMkc8AZxVWFckd+grQgQAg47jrzWiehLRagTGM+xq/EoGOvSq0K8DjpV6JTkHp+VUSyxEOn+eatxgZBqCJSAOKtxDgcUmSSxrkCrCDpz2qNBjFTqOlSIkQYxUoFMUdKlUdKQDl7fhTwKbingdPekAuKUUfypcdPSgEGKMdKXFGOaB7iYORS4pQKUCk2XFAB7dKXH5UoHtS4qGbJBijHSnYopFpBilHGaMUoFIpBS460evrS0AFL60ccUUIAH50Z5o96KYgPHaignpRTAM0HNGaTPWmAUdaKM9KVgFzRmkz0ozRYBc0Z/wpKKLAL6+lApKXNFgFzRmkzRRYBRQO1J+lLn8KAFopKKAsLnFLmm5paQhfSgUn/wBalzQAtFJzRmgBaKKKAFzRnrSUZosKwtHSjNFKwC5ozSUUCFoozRmgAoNFGaAAUuaSigBaKM0gosAtFFFFgClpKKVgD1pe9JRmgQoo4pKWgAoozR/WiwBRRmgUAFFFFABRRRQAUUUUAFGaKKACiiigAooooAWijPSjOaBBRRRQAtAozR1oAKKKKACik9qWgAooooEFFFFABSik7Ufjz/WgBfSjNJxRQAo/KiiigAx0ooooAKKKKACiiigAoo/nRj3oAKKKKACiiigA9KTFLRigBKKX2pKB3A0UfhRQFwoPSiincQlLiijFAxKKXFGKAEoH6Uv86SgAooooAKKKKADGaKKKADHSkxS0UXATFFLSUXGFFLzSfWmAUUYoxQAUUUY60AFFFFABRRRQAUYoooATHSjHFLRQAlFLjpR/SgBKMUtHvQAlFLiigAFJS0nWgAopaMf0oC4lFL/jSUDCiiigAooo5oAKMUUUCCjHWiigAxRiiigApOlLijFABmkx70uKMUAJjFLRRmgBKXFIKM/jQAY6ewpaKBzQAmKXH+cUAdKKACjpRRQAhox1/KlxRQAY6Ud6MfpRQAg/+vS4oooAMUYoxRQAUmKWigBP6UuKMUUwDFJS4xRikAlLikxRgGgBeaTHWgjNLigBMdaKWjFMBKBS+tJ7UAFBox0ooADR6UUUAHpSYpcdaMUAJRS+lFACUUv8qQ8+9ABRRiigA7ZoFFFABRiiigAx9KMUEdKKAE//AF0uKKMUAA/+vR6UYooAMUdqPSjv9KACjpRiigAP8qSlxRigQYpKWj8aADmgUYooAPwoHWj/APXRii4CUpo5pKACgDr6UuPzpKLgFFL6flSUwYUY60UUCCjHWiigAooooAKOKMdaP60AFFFFACUGlooATtRj8KX/APXSc/nQAYo/Wj0o70AFGKP5UGgA5ooFHpQO4etFHpQKAF5pMUH+dHrQAY6UUUUAFFFFAAR0oNHqKKAClpKKACiiigAoFGKKACjGKKKADNFGOKKACjHSiigA6Gg0UUAFFFGKAEpe1FAoADj0oxRij3oBhjNJS4/SjFAg4pKWjGe9ACUf40vFGOtACUHtRS9aAEx0ooo70AFFL/PrSYo3ATHvS9qWkoAT60tGOtGPyoATFL60UUAGKMUfjRigLCUUYpeKAsJjpS4ozRigBKKXFJj3oABTgM4pBmpEUkjik2Gw6NOnFTKOOnFIo6cVJ0xWbdyGwpaKBQkSLSUtFMApKWkoAOOtLRSCgA7UelH6UtACdKPSig0AHpS0gooAWkNLSZoAWkxQaMUAFGM0UUrDInQEHFVWQgmr554qGRAQT2oTsUmUiDSYFPZSCeKZitU7lBijtR/jQKYBRRmigAx0ooopsAoNFFABmijrRwTQIDSZpaTFABRRR680AA70frR2ooAKKKM/rQAUUZooAKD2oo5oADmiijNMA4pKKXFACfhQaKKAA9+1FFGKAE/lRRiigAoNH6UUCuFFFH8qYBjpRmjmkoAKKDQT19hQAUmKWkxQAH6mg0elH40AHrzRQaKAD3oH1zQaKBAaKKCc07gGKMUZ/CjNFgCiigdqGMP8aKPQUUgCijBooBBRmij+lAw9aKM0CgAo/lRij1oAMUUUZoAPSg0Yz7UdqAD09qOOaO9Hp70AHrRS46UUAJRQaMUAFHFGKMe3agA60Ufzox1oAKDS8UlAAO1GKKPwoAOaTFL60UAJ/SilP5UmPyoAKO1FGaAExS0Y60UAB70mKOtBH+fWgAoNFGM0AH0o6fWgjrRTASg0tJ70AIQeOKX0o9qKADFGKBxR/n8KADHWg/pR2o7/AEoAQjk0H9KXrSUAHWkI60tGeKLgJjpRzRilP5UXAT+tLj2pMUCmAcAUmKWigBKO1GKP8+tABSdPpS/0pKAD+dBz/n0o+tHOBQgE+vWjFKaSmAGjrRRQAfjSY5paD25oAQ0mBj1pxpKACkxSmigBP60dv0oxRigAx09qB/8AWoFB/KgAGM0mKX/CkoAKCOvvS46+lJQAY6/nSd/c0veimAlHf6UvU0mPx/SkAUcfSj+dFMA6AUhGcetLjpRQAhoNL/OkoAMdKP50UYoACBk0mKXvR+lACfhSetOI9KQ0CE/nQec/lS0nSgLBijmjuKPWgAxSYBxxilNB70AJSYIpw6fSk70XATHtxRR/OloAT17UH8hRjFB4/Gi4hKTg/SnYoOPxoAT8KD+lGPeigBD+n6UnWnYpP5UAJ6e1BpeMGkwc0CYdaTH50v8AKg0AIeMUHtS+nrSd6aAQ9RxRjvjrS4z+HFIfwoAAPyAoA64o9Ox/nRQIKSg9gfpRQAH/AOvRgemaKD1p3AM03PrTuf8APrR/n/GgBpopaTANAXDHSj17ZoNFAhMYHSjnNL68UnegApMfiP0pcnP+fyoOOaAE9OlGPzo/TpRxRYBKOn50vpSU2AGm4x/KnfjTT060AMbnNRtkZqQjrTGOelUgIz34qJhjPH9KlJOTUb4wKaBldz1+oqvJjBx2H9easPkk896ryY57irQitIMg89Pz/CqsuOR6cVakyTgDP61Vl4zxz19Pzq0JlKX3+v45qq+cdfr/AIValBB4HOD0/pVRyAT1/wDr+9aIhkDfe6Ec1EckkEkDPBzUjA5PPTP+c+tRt17d/wDIqkSIAR19uvenpnj8qYBngH8aegGQcHtQJFuIcn0/T6VZVTtHQfjVaMnjntz27VaXlRzUso+dqKKK8M3CiiigAooooAKKKKACiiigAooooAKKKKACiiigAooooAKKKKACiiigAooPWigAoooFABijFFFAAKXB496KXmkOwdKXFIPend/8igpADTvwpAORxTgAM5/CkWkwHP8AnNO/rj86THPp1p2QCD+fSkWkLj0HTn607BOR26/SjAAPccUqgHGQalstIcAeOg5x680oAGTznFA6n8/T8KeOSRgAc+9S2UkCjg4Az9f8adj6En8fypB2Puc59aeMZxn/APXUstCDABx249acM8jHsaQLyM8duD/SngYxnrgD1pNlJCYwCSOemRzzinbSe4GefT8qTHX0HenDIPX3xweKlspIO5yQefr+VOAJ746UncZHpz7U4AeuAfXnn/A0FJCjAJyM8Z4H5CnY4HPP+e9NAIH169804cgEgenpUvctBgE+wxn1p4B5IPIOOcU0ZPsQfXOfxp4wckcD0qWUgA4Hb171IoI79QKYuDkfU+9PHBzjJ/yKl9ikOUYz1wP1p6j2Oc/XvTVB5Oemffj/AOtTlIyOeOec1DLRIg9vYd+Pap0Gcc+304zUSA+vf3/QVMvBBwcHjuKhlrQmQZA59qkAPfpnJ/xpijOMdv1p6gEnimkMkGCBz1Gc4/KnqMnJz7imqMZGD/LtThyQAeTnr061SQx68Y/KnjnHHQA+lMHBznrn/Ip6+4z7YpjTJARjjt29qkGTnB/w+majXBwDx0PPFPGMnB70hkyDGBnOfT9asRAZHHA4wKroOoI9atwjgY9RQlqNsvQgAAntVlOASPXPqarRHAHPvxxU6nIB6DOT+VdEdjGTJwSCOOafngc9Pp6VCp4HHHr7+3tTsnAxnjk1otDFq5IWPP0HvSgg559vb/8AVUYOcknnoKAcAZ+nHFUmQ0TA8AEjtRnBPH/1xTAcA+nU0u7HB5B/zx707kj+p5OenvzQSCcjPt2xTAevI+p4/KjPA/PrSE1cfnt3GKQk4Hp196aWBJ6c8CgHkjPPPHH+cUCsOJwB+eKjJ65PPB460Egcn19cU0kAHPbt1/WgLCEA5JOOh9RSEA5wecZ/ClJwD69aMEkdc88+3elcLWEAOAOvXHXpnvTtuOhyDgn88U4AZH/6qdjH/wCrt24oTJY0Kcc9+D/jTGBIIyOpPbHtUuMk8egppHTj688YqiWiuykZz14OcY/WonTOePw61adRk84x/L8KhKEg/p+fWqQrWKbJjIAzyexH51A6HJHcHB4xz/jV9kGDj8zUDIOMjimhNFB4weo55NQOmRyOefpV9ozx+v8AhULJkk4/IY7UyWjOaPBPA5I49KgdDySeMkccVovHnqOP58VC0fXjkcfj/hVCt3Mt4wABjGOeTmonjHPp2HPWtF4unp2qExdc/r6UmgsUDET26c9MUwocHg8n6+1XzEcgDuPrTfKyenPTpStqKxRMWRjsOKPKJHIA7j1/Orvlfhg/UY70nlcjA4HftimkDRUEYOT6c9PzpRGcjHt78VbCZPTg+1KIQRjOOc49qpIVrFQRn0x+FPEXIOB3H+Bq0IuBwOcDp29qcsWe2enWmkS2QeUeMD8e/wClPCcDAHGM/wBan8o84z36Y/pT1TOOPT/OKpRAjROfbjrVlFIA/wA8ULFjHA54NWY48AZ9uv8AKqSExYkAA596uxJkjP8An6VHFGTjg9/Qf5FXYk6Aj29Oaaj2JaViWFBkcZwelaMKkEen0zVeFMEkjPAx/wDqq/EhHY9gPWtEtDNtFiJORkdcD07VdiXGPwqCJDgetXIl4HtnmgknjHA75/SrUa9OahjXOPwOParKLjFJkkqDp+X/ANap1FRqMYqZQBikxD1FSgfnTAM4qQDrSYDlH6Gn46U0CnYpAKPpS/hwaMfgKUdaAEx+dOA96Mc/WlFJlJAO1KB+FFOA/nUs1SExmnAf40ClxSZogx0oAP6UuOaMdqTLQUooFGKQxcYpR+tAo/WgA9aKKKYBmgnFGaM0xBnHeg0lJmmAuaD3pDRmgAz1ooozQAUGkzRTsMUUCkz1ozRYQtGaOtGaLALSim0Z/CkA4fyozTc0tAC0tJmjNKwC0uabS5osAuc0elIKXNAhc4/OgUlFAC5ozSZ/wpc0gFopM0UALS5puaWgQuaM0lFAC0ZozQDmgA/GlzTaKVgHZoNJmgZosIWiiigApc0lFAC0UlLmgAoozRRYAooooAO9A7c0UUALR/Kk70ZoELRRzmiiwBxRRRmgAooopWAKKKKACiiigAooooAKKKKAFopO9LQIKKKKAF/Ciko/lQAtFFFABRRRQIPWj2ooGaAF9KP8/jR6Uf8A66AD0oo4FFABRRRQAUUUUABopcUlAAKWiigQlLRSGgBcUYpKXFABijFIKXvQFxKKXFJ/KgYUUuKSgAxRjpRRQAY6cUmKWigBMdaKWjFACUUD6UUXAKKMYopgBpKWj0oAMUlKaTH+FABRS4pKBhRS4pKACiiigAxRRRQAUd6KKAExRilooGJijHFLRQAnpRiiimAUUYo7UAFFGKKACiiigAooxRQAUcUUUAFBoooAKKKMUAFGKKKACj/OaKKAADNFBooAP8aSlNGKAEox09aXFJQAUUf/AKqKACiiigAopaSgAx0ooooAKKKKACiiigBf/wBdGKKKAExS/wBKP5CigAxSUvWigA/lRiiigA/woxRRQAYoxR/n0ooASlxRj3ooAMUh7UuKMUAJjmilxRj9aAEopcUYoASj9KMUUAGKMUUD9aAD3pMAUtFABijFFFABik6UuKMUAJRjpS4o70wEo9aX8KMUgEooNBpgGKOpoooAMUYoFFAAQKTGKWjFACUYpTRigBOlFLij0oAT+tFLg0Y/zigBKMfpRiigAopeOaSgAooooAP5UUUUAFGKKKBBRRRQAY/xooo5oABQegoxRmgAooPSjFFwDFHcUYo6UAJRS9aD1ouISiil9aLgJRS+npRii4CUUUUwCiiigApe/wCHekooAMUdc0UUAGKT0paM0AJ/jRS0mPxoAD+dHpS80n9aACg0vp70mKBgKOf60UCgBaTpRxzRigAooxRQAf40CiigAooxR6UAGKKKKACiiigAooooAKKKKACiiigAoo9KKAAdqBRRQAUUGigQUcf0ooNAAKOlFAzzQAUUY6UUABoFFFABikpaP50AGKMf40UUAH+fSkx1paKAE70tHH9KKAExRR3pTQAnejvS/lRQAlFGKKADFFFFABjpRiil9PWgAAyRU6DAFMQZINWFFRJktigYFOFAoqUS2LRzSClpiENH86Xmk4oAWk70tFABSY60elAoAPSij0peKAEo9aOaP60AGaKP88Uf55oAXtRRSfqKAFoopPSgA/xoo69qWgBPakYZBHFL+VB5pMCrIgGarsME1dcdaqSDBqoPoaJkeOlFBox0rQYUUUU0AdaKKP170AFBoNJ3oAKKWkJ/CgQUZ56+1FHfvQADvRmj1pMnNMBc9aB+lFJQAtFJmjOe1ABxxS0DFJQAtHp7UlLQAZo9aKQ0AH9aP6UUH07UAHQ0UUdKAAUf5xRwaKAD0pDS/wAqSgTDHSjHeg/zop2ADSUuKSgBaSiigAooooAKTNLSf40ALSe1HbpRmgQevpQaPWigAooo7UABo7UUU7gH8qDR60etDAOaKOaKBhRRR0pAFHTHegjrRQAGijriigYdM0Y6UZ5FH9aADFGKX8D6UnpQAtJ25o/rS0AIaP8AClHbiigBBSj+VHpRQAUUUUAGOtHpQKM80AAHSiiigAxRz+NGOlFCAKQ0tGOnNA7CYoo9O1LQFhKKB+dHoaAsGPeg0Yo70CDFJ3paD/KgdhOfrQRS460lAWCkP8qU5o9PagQYoxR6+9FACfzopaTHSgA9KOtBHWimAYo9O1HHHFGKAEx1oNGKKAD07dqMfpR1NHegA5oP8qKKADFJRnOKX8KAEx796KKKAENGKWk//XQAY/Sg0Ue1AB0o9KBSen50wCjtQeMUUAFJzRS/hQAnvRS0goADx+dJS0hxTBBR/wDXoNGDQAlLjp70UYoATHSil4oNACfhR60uKT+lABSelLzRQAlBpTRxQAlGOlGPyooAQjr7UY6Uo4pMUAGKKMUD+dABjrSYpaT/ADzQAfjQaP5Gg0bgA7UmPWlI6UZzTATn9aD29qXFHpQAlGOaM9KM/lQAc80elFBycf54oEhKKX0/Kj8KAYnrxSHp9aX17Udc0AJRS+mKTNABRQaMflQAUlKO3+c0hzQAYpD0/wAilxxRjigBMfh3o/8ArUGgfoaBAaTHINKB+tJg9KADijrgf/Xo6/yoPNABik5xS/5/GkoEJwc0e/oKWkHGKADp/OjsOxooPSgBP6c0h79qXGKD1piE6Y9KMdKU9qQe1ABjGeKCPSg9T2oI6elACf0ooP8A9agDpTAM0h60p70nXJoEGaDxR3pMHmgA/pRjrRxRzxQAneil79aPSgLielA4pT274/WmmhAGCc80vaj8KTnjn+tMAGMetIec/nS5/Sj0oAaQfXikPHSnHBpp6evamA0+9RsevPPNPJzj6Uw8ZoQXI2HBP4VExPP1qVuwqFuv+etUDIXzz/nFV5ODVhzkGq8h4POatCK8gPFVZSD39f8A9VWZDkH6VVl/I4/+vxVoTKUwPJB4JH/16qSAkZJx0z2/OrcvU8+uKqScZ79/T8a0RmyBh1yOPSoieef8/wD16mYZH4/561CTjJ749P1qkS2IRjBz/Tj6etSR5IA9x/8AqqI9B2/CpUAwD3Hf2piTLMYPrjj6Yq0p+UcfqKqoQegPHp+fWrAAI6UirnzzRRRXgnQFFFFABRRRQAUUUUAFFFFABRRRQAUUUUAFFFFABRRRQAUUUUAFAooIxQADrRRRQAHrS0najvQAUtJmloAUDNGMgc0f/qpaRSVwxilAz2oA/OigpIX0604DP0pAP6U4ZGealmiQuO3rT1xggnjrj/Gm4xg4/wD1U4c5BH0NJlodjrjn/wDXxTgCBkH2pAOB+XTmnKAADnjIPr+lTcpDgM45Pbj9fypQMe/fFAyT14H+f0p3fA75/l2xUtloVRjqPTH/AOqlxwD1z/n8qQH29/Q/jTuCenp1596RSF4PQ8H0/LilGeQRnoKFAyOMdaUAjgd8/wCNSWg5z9M8UvJAx15H+cUcEc84460vHbP8sikNIU5wTyO3vSqO/Pf60gOQOOv4Uufw/T9TSKQoOAeefXr9fwp2OCCOOcYpByTxz1p35nofw70mUKAOMHAPt2+opwxk4PA5x/ntTQDxgnkACnA4zzxz/wDqqGUhQQTke9PAORn/AD+vSmqBgcev1P408AH26ntSZaFXg9R6DipAAccen4celNHPHY8Z4zmngYBwe+fX8qhlIlUYxxkc4+vTmp1GMdefz/8A11CvT8cDvxU6kHH41NiiQc89f8/zqUd8g8n6e1RqOCeffP0qQYJz69f/ANdUhkgwSMnt6k0uMd+574puc455/pTx3P1z2qrFDscDp/Ufh6U9c5x9aYMnoOegpwOQDyT2/wAmgB/Iz/n8v8KlAJAyR69Mc+9RoDnjpUijJ69SPekNE8Z4z1HT079qtQgZGPpzVVOcAHgcelXIRjHHFNLUbZbToMDsPaplxjqe9QqcAYOeg/xqQEYJPB/zg1sjGWpMCAD+XbFLnAOD0qEMcZwD2z1pxPIHT0q0QSg8Dkde9LknJOM81CD1wR2pVJH6ZzTTIZMD1APJ5oyfXn16VFuHIzTg3GT6euKdySTIIxnJH40bgR1x64qIHDEg8dOeefY+lBck9eOnpx3ouFiXdkHHHXv/AIUmRg5OO/v0/lUZbBIHbnHSjcCc49+nWhsViUnkjtx0puRknOe/+TTcjIAx1HbNKSR/nj3FK47Ds9Cccc05QSTgf0NRgnIOc5/zzUi4ycentQ2S0PUA8H1PtTgAc/gP/wBVIhPGe34fzpwGCAc8Z6UIloQDB7YpCOv0x04qTAAHGe3Y0HBP+RVIkhZSfzz/AI1GVznnrx7ZqyQOoyOPpTCnt044FWhFVkHIxwSB9aidCc8VcKAEknnqfWmMgOcDt/k01oQ/IpMnUEYx0+ue1RNHyeOnUf4VeaLjI9wDUDoc4I56/TtVIl6FF0OD6Dt/nvVdo85yD+XbtxWiyDnoc5P+GahaIEk8+ppkmc0QGcDsT07e/vULRdSR7/8A1sVotGMnJ46f/qqMxEjOP0zTtcZntEeeODx6U0xH0zzj05rQ8rI4Hv6Ck8o8HGTx2/pRYGyh5J4OOP0xmjyTzxzzx74+tXxDyfUc0eVngjv9P5U0hNlHycgZHtjpQYc44P0HHFX/ACj+Xtml8k56j19KaRLKPlYIA+g4JpwiwPr7Yq75PIyDk4PQH8Oad5ODjHB6cY/CqSfUloorEc89uKkWLqMEdPb61cEWccY/z0qQQccjnrnnj9atK4PTUqJEcDg846fr+NWI4gQOMAnGB/8AWqwkHB/w7VPHDwOMYPHY+lUkS2RRQ4xwcZwB6VdijII9v89ackWByPerSR+3OfTtT2JctBYY8YOOOv4+9Xo055H+e1MiizjjgYHfirkceCD/APX4pkNj406YHX8atomMUyNMY4x2q0i9OOtTclkka89PSrCL09etRxrjGasIOlJkj1Gce1SqMYpqDOKlUdKQMcowB2p4FIop4ApMBeacKQDp3pwHIoAAPzpQPwoA6Uox6UAGKWjvSjtSZaDHWnAUlKPrUs1QopaKWky0J3NLRiikWg+tKB0o9KP6GkMXFFHFFMQnP50Enml4pKYBRRSGmAf0ooo/lQMOaM9aTNFABRRSGgAzR29KDRnpTAXiikzRnmiwhaM5pMnpS5osAfjS803PSlz0osAoopM9KM0WGO6UUgoNIQ7NGaaOvpS0AKD2pc0maBSsAue9ApKKLBYdmkz7UZ6etAosKwtLmm9c0tAC5opKM4pALmlpBRmgBaKQUZ6UALRRRQAtFJS+tAgoozRmgBaKSigBaXNNpc0hBS5pM0UAH60daKPWgBaKKKACiiigAooozigQuKKT/wCuKXNAAPyooooAKKKKQBRRRQAUUUUAFFFFAC0UDtRQIKKKKAFxQKPwpOKAFFFJzS0AFFFFAgpfpSUd+KAFoFHX6UY60AFFFFABRRS4oAQUClpOlAgpaKKACiiigAooooAKKKKACiiigAooooAPxozRRQAmOaKWigBKKP1ooGFFFFABRiiigAxSY60tA/U0AJRS+lHrigBKKKKACiiimAYoxRRQAYpKXHFJ6/nQMKKX0pKACilxSUAFFFFABRRRQAUUUY6UAFFFFABijHvRRQAn+NLzRRQAmKMUuKMUAJijHSl//VRjNFwExiijH9aWi4xKKP5UcnNMAoo4+tFABRRRigAooooAKDRRQAUY/KiigAox/hRRQAc0Hv3o60UAJjmjHWlNHPNACUtFFACYo9qXp/nFFACUvaiigA//AF0UUDmgAooooAMGiiigAooooAKKMUUAFFFFABRRRQAUUUUAGKKKKACiiigAooooAPpRzRRQAlKOtGOaKAEo9aWjFACUUvP5UUAJRRRQAYooxRxQAfpRiijFABik+tLRQAmKWiigLCUuKKKYCUUuKO9IBKKKWgBKKXFGOKAEox1oopgFBpcUhoAMUYoooAKPWg0UAJzRS0YoATFHNLj/ABooEJiil4o/nQAlFLR68UAJRjFFFABiiiigBM9OlLRRQAHpRzzRRQG4CjHP1oxzQelACUpozQaGFg70lLj86KAEpR1o65oHWgBKKKKLgFFFFFwsFFFFMAoFFFJgFGKKOlMAo6UUUAFH9KKKVwENLR60etFwENHalxRTAQUdcflS+nFGKVwEx/nFH9aXNFMBKM0GloASjFH8qKACj1o6UelAB3ooooAKKBRQACij9aO9AMKMUtJ/kdaBBijjFGKMUAH86U96TFLQAgzmjHX3oNHrQACijNLQAlGOtLxSUAFGKMUvFACUUYpeaAEoxR6UGgAooxR/jQAUUUfWgA70n9aWjFACUcUvWjHIoAMU5RkimipIxkik3YCRBwKmAwKaop4rPchsKBRRTJFpPSlooASlpDRQAZpaTPNFAB/OjFHpR/jQAdKWk7c0d6ADv3o69qPeigA60Zo9aWgBKKOPzoFABR/hR6UtACfyooNLQAneg0UUAhjDIqrIOTVwgGq0g60k7MtFYjFJSsMGkrZalBRQKKdgCjPNGKKAD/GiikzQAtJRRQIKKO1FAB2o5FJmlzTAQ0tFJQAUUUZ6UAHtQD7c0Dt+dL+NACZ/SgUp70ZoASj170HPPSloASlxSfpRQAUcfnQaKAA0cntR1o9aAEozRiigAooz09qKYgoxSUtABmjpR170lABRRRQAUUUUCENLRSYoAKKKO/vQAUUUUAH86KKKYBRmjFApAA7UUUUAFH86P50UDAdfSjpQaPSgELijijHWkxQMPzzS880n9KU0AFGP1FGOaKACiijsKAAUdaKWgBPy70UYOaKLgFL60YpKACil/pSUgClNH40n64osMKKU0ZoASil4pKACig0U7gFJ2pcUYoYCGj1NLSdM+lABRR+PNGOtABijHT0oooATH60d6XFBAoEJR2ozR6etABmk65pcdKMDFACdKKXNFACY6UY9qXFGKYCUY5oooATvR680p/z70etACdKKPSigAPSg0HpRQAfjQetBo9CaAEoxRSYpgBopfWkNABQaKKAD/OaSl5+lHegBOMGiij9aB2E/nRjpS0mPegQE/wAqKMGiiwBQP0FFFCBCdSKMf1pT270evFFwEpOhp1IaYAf5c0UHkigjg9qAEx0opcUYoASjHSlxSUAIaMCl9Pek/nQAUUUUAFFBANH+NACUEUvce9IR7UAIQeKMYp1JmgBPTjpRxxRilo3FcSg0c8+lB780xgKSlx+VGKBCUGlIPFJQAY6UetJ6jrS0BcOlHr6UZ4o/WgBKD1PFL+HFIc0AGKQ0tFACY96DRjpQR1oAKQc5paKAEoo//VQPrQIMUn4ZozmihAHakpRRQAnNHNGOaDQISkx19acf/r0nc8cUAJx/SjPFHTHf9aD1pgBo9eKKMe/WgQhH40mOnNOpDx19aAEx1oOfaj+tHr+NACH+dBHSlIPFHTNMBMdKBR09aMUCEpCSccUtJ/8AXoAD70fhR6elBzmiwB0zR0Ao9qTv9eabEFGc5ooJyKAE4+v86COlBxz60dc0BcOc+1B70Htz3o/WgBOlNI7d6ceg96aeuPfNMLjG5+lMbgGnnODxUZOc00BGw/xqNv8AGpGqNiDnimBA2efy/Cq7jPHP+f8APSrDk884x+FV3x17c/5zWiAryg8/j04qq5Oe3rjHFWpOc+vPeqkg4IOT/OqRLKcoz6e+KpyEZ646irkvOevI+nNU3yCRn1rRENkLHnA6ce/44qFuecYOR3wcelTE4yetQuM5znrnA4qyGxvXB6HOO4NSR9T6f1qPtjHJODT04IJPTA+vtQItRcA9ffHarSg7RzVaIknGP61ZBGOlJlHzxRRRXgnSFFFFABRRRQAUUUUAFFFFABRRRQAUUUUAFFFFABRRRQAUUUUAFFLxSUAFA60UUAH4UfjS5ooAKUUetGMfhQUg6ilHBpO2KdjGaQ0KP896Xjt7UgyM/lmndAPWkaIB7d6dgcccfrSDrThk5wPf0pMpC8kA4p4AyM/WkXoeOetKAeMj0A9vapLQ7pk+mRnNOAHHPpn29sUg6E9xn2p4wBz15qWUKvGSRz9OfanAkehP+c9Kb3IFPGCTyO/rUspAAQOR6f54pR/hz0oUE5I7c4//AF0qgdD/AJ/KpKFJIPHToe9OGAAT7jik46Y6cf8A66UEnOeB1/pSZaAEEZx/n60p5zjJI5pMckDv/k0vTn6f/W4oGhSB19PfFOGR0wB3700gYznHH5/SlyeceuR25pFDsD36cfSnYAH6Cmg5yc8Zzz+tPBPOcc89B6UmWhQMAc+hGOn0pR1JPJyM9qQAEEdB29MU4YHf39fzqWUkO4B5OD2704AYJGevNNBPTOTz+XtTgAQO/wClSUh4wTzj/J6mnqTnkDgZ/wAaZnoevrzUqdyOufr35rNlIlTBx7/lU6DOM8Djp61CoOQenpUyA8DHT8eKErlEi8ED/OfSpFJGRn2/wpg4GeM5P1qQDB4JH/6+2KpIdxwAxyO/r+dPAIx6fWmgAjqOM96cCDjJ9c9elNILjuODyc4IpwJOB1wD2/zzTQM8+36U5TyMHHX+VOw0yRccdR+tSLjA5znntTFGMH/Dn0wKkUZIz0/LmpGiwnB6HP5frVyL1A5GPeqcYBIPcf5/OriYGefarigbLCkA9/YdaeCDnPTt2FQAgkY9fXFSBuRyOw/rmtEZMfuHOe4pc5HI/l0qPIJJ7jOT0/Klycjkd6olokzxwOPXpk/hRuz29e9Rg4PPU9O3P1oLAY6n279fammS0S7zz2/z/Kl38enqM/piot2AT6gcde9ID1x9fTt607isTZIyc4GPxpN3I54/Ko9xABz689aaW6c8c9/5UBYmBIyATxx+tKGwD7dxUJc5HrRuBIx7VNwsT7gSM5J/zyRTt3OCOP61AGGeevQ+/NO3E4xxgfSi4WJx7H35/rTwQPU9ufzFRIc5OQM4/wAmplIJHp1/CgTRKuCPXg9Dmnrg4z9KjTkDHHXpz9c+1SgZAB+nHGe+KEZtDgCMHjnP5fjS4I9/60oHIwMdqcAeuOhyR1q0Sxm0ZJz/AProKnnHfnuM1LtwB3/WlI4+nFWZtlYpke/6U1kI+ufrVkoOuOMDrTShyDj6VSE2VWTPbHfqKhaPk8jJPPfn3q8UyenI7459fSomj6nHr2qluQyg0YOePaomjwCMf/q7VoGLPT6nHpUZjBz9MVSVwM9osgkDnvimNEQCcdh/nmtDyj1x/wDXprRDAyM8gU0gbM8xDjjr9RSeVgnIA7VeMXJ4OB+dJ5We2O3TOfrmqsTcpCI8/n60CHBHGSccn0q8Is8Ht/KlEROOOnP/ANb6VSRNykIDjpnsPWlEOPoM+9XvKPP5GnCH26e1NITZREJwBj09cYpwhJB44/z0zV8Q9Bg456U4Q9BjHr70xNlJYMkfTI+tSLB1OM55q4sIA6flzUqwdABxj0qkyOYprb4AJH9anWHoMdfQ4q0sI4wOKlWLGOOn5fSq8xNldIT/AJ5q1FEBg4/lUiRdOORxVhIsEYoJvoMji5HHT8KtRoAOBz0pY48HoP8A61WFTocAdKRLYInQ464qyiA446flSIh4PUfrU6rj+dSxDkXpUygcd+lNA6cVMo6UhCqDxUoH50gFPAoEKBxTwKQDpTwKQ7hjr2pw6CkApcUBcBzj0pcdKMdKXFIAxQKBSj+lDKQoo9KB/KgfrUs1iKKWkpfSkzVC+n50UetAz/WkUH86WkFLnFAwpc0mfyooAKKM0mfwpgLSUUUxBRRjpSdOlAwooozQIQ0HvRSUxi0gPaj8eaAetAgoHb1oozQAZ5o70UUALmjOKQUvbrQAZ6UuaT+dGffFFgFpc03PvS5oGLnk0Ckpc0WEGfxpc0hoFFgFzS5puaXNIBRRnFJRQA7/ABoHGKTNHNAMWlpoNKO1KwmLRmkz0oGePSgBwozSelFIBaKSlzQAdKWkzQKAFopBRmgB2aM03NLmgQuaKSigLC0tNyaWgBc5opM0uaBBS5pKKLAA60tJS5pWAKKPxooAKKKKBWDvS0lKKADNFAooAKKKKLAFFFFIAooooAMUtFFAgooooAKKP50o6UAFFFFABRRRQIPw96X1o9aOlABRRRQAUUUUAFFFFFwCjFHvS0CDvRRRQAUUUUAFFFFABRRRQAUUUUAFFFFABRRRQAUUUUAFJS0UAFJS0UAIaKXmk696ACilooGJRRRQAcUUUUAJijFKaO/vQAlFLikoHcKKKKdxBRiiii4BjpQBiijHWi4BijFFFABikpf50GgYlHWlxSUAFFLikoAKKKKACiiigAooooAKKKKACiiigAooooATFHtS0lMYCijFLzQAhopcD1pKACij1ooAKKKKAA0UUUAFFFFABR070Ud6ACiiigAo6UUUAFGKKKACiiigAooooAKKKKACjmijtQAUYxR0peaAEoxR6UZoABR/+qiloEJS8Uf59aQf1oAKKP50YoGFFH/6qKACiijj1oAKKPWj/GgAoooxQAUUUUAFFFFABRiiigAxmj/9VFFACYpaMUUAJiloooATFGaWjHFACYox0pcUfyoASilpKACiijFAB6etH6UYooAKKWkoAPWkxSjvRjFAB+FFFJj3oAXFJS4ooASil/8A1UfjQAlFGPeigAo7daKKYgo45o/KigApKX1o6d6AExRS496KAExiiiigA/CiiigAoooPSgAx1oxRRQAdaSiigAopfxpKACiiigAo5paSgApTSUtFwEoooBxQAUUUUAFFFFABRRRQAHiiil6UAJ070UUUAFFFFABRRRQAUUUUAJ/Kl/LrRjpRRcBPWlxRjpRimAmKOlLRigBDx9aMUelHbpQIP/10YpaKACk9KMUvvQAn86WijtQAdaOw9uaBzRQAYoxRRjmlYAooopgGKMdKKKACkNLRQAUlLRQAmKO31o7UtACYoPGaMUetABR0oooAKDRRQAf40UUUAKACamjFRDjFTR9RUSEyZeBS0g6UtSjMMUtJkUuaYBSdBS0UAJ/+qloooATrQKKWgBPpS0n86KADFH0xR+lLQAlLSYo/OgAox196MUtACf0paT8aPWgApe1JijFAB1ozR6UUAFFHb/CjFACH9P51DIOtTkZqJx1qdmUtymwwTTPWpHHJqPpW0di0woo5oqgDNGKKKAE70UppD+tAgooxRQAUUUHNMApKBS5oAPwo70cUZoASlxRSZ5oABR/OgD8c0H8aAFopKM9KACij+dLQAfhRRRQAho9fWg0dutAAKKM9aKAD+VJjpS/jSUCCg9KKKAA9KKMUH+VMAI/EUYopMdfrQAUDP4dKKXHSgBKKBRQAUUUYoEJ+lLRRigBKMdKXFJ6UAFBHT2oxQKAD+dFGKMUDCijFBoAKPT8qXiigBP50tGOtKKBiUUY60ooASj+lFLigBKMdKXFHFFwQYoxR0x9aMUXAB1pP0pTRikgDFGKMH2ooADj1oo6UUAg5ooI60UDDH40YoooAM47UZIoooADR3oo60AJS/l9aPT8qSgAx1ooopgJ/jRil/TvRQAn/AOugjrS0c0AJQfyoNGKAA0elBooEFJS/yo6CgBD2oo96MUAHajtRiimAmM/SilxSdKADmilxSfzoATvS0dM0GgA/wpKU0lABijNFGOvNABRR6+lFABikpaSgAxRxR0/lSf5NMAoo/lR+lABg0cfjRRQAlGP8KXFIPrQAYo7UYPNFABSGlzSH8qAFozSc80vagBKKKMUAA/8A10mKWk5pgHP0ooo9KLAGKKBQfagBKPpSmkzj60AH88UYxS4pP1oAMdKT0peKKAE/Oj19qOlGaAD8KMUUdaAEoox0ooAMDFJ0+lLRQITHTilxR+NJn9aYAR/KjjjtRmgUAGKMUY680HnNACdzRR0oxQAHpQMUZ70YoAM9KD1ozSUAL60nr9KU0metABxSetL60nvQAUUUCgBCPyox/hS+vrSdaBCYpfSjtR65oASj0ox60GgBKPUUtGMetAhDQaKKAEP1pCODS0elACfhQaMUUxCEfl/OjGaO9B4oASjnn1pT2oPegQhx9DRR/wDqoxTASijPSjnnsKADrSdM0dPoaMdqEAmPajv1paQ5zTEHFJ3pc0fyoAQ80dqQ0fSgAx3+nagUccelFAB6f/WpD0PpQSOfWg5oAPbvTTwKce9NPQ+nFMBjZ4574qM98CnnPFMOACPUfpTAjbOPwqJsgH6VKe/+eKhY9c1SERMSQTj39qryHGeeetTsccd6gfIJI96tBcrSdTVSQAE89AR6Vbk7/wD6qqTck469u9WiWU5TkEDp9O/aqj8+/I9qtyggk9+T6fTiqbYPGOv860Rm2RMQDyOPeomJOQPU9AakfOMemcComJOPz9f51SJbGk4AJzzx+vvT0xnOPTHemkYAzz3/AP109M5GeeM8UxFlMAD9asqBj/8AVVaPOMAdf8PWrCnjoPzqWUfPVFFFeCdQUUUUAFFFFABRRRQAUUUUAFFFFABRRRQAUUUUAFFFFABRRRQAUUUc0AFFGKO9AC+lFFLjpQAfyox0oHBpe9ItIAMf404D2pB1FOwQBSZSQmP89qd+FA65p2MEHFItIB7D0p2OT6fnSDJycelOAIBP+TSKQ4AenPHWngYPXnn/AD9aauDnnkfj9aeBggkgdcelQWgweMZHTjrTx3GOPzpOTgY4FOAAAJ5+gxSGkGASeOvOacARz1/n70ADsMnn3pwHJz6jHv8Ah61Ny0gzyeef8/rS4Bx27n86MHg/5/A0oPQkY4A7evNIpIXGASfSjAx1z0oBGDz7k0dCeM/1/KlqUgHGe/PHrS8ZPtnANJjGSc/Uj8aUjnOe3/16GNAQB3yD9R/k04DJ9hzjr7gUnBPXt+dKucg8Yz9OOnWkWh3JBGOwyOlLweMkHgfhSLjBIJ9RTgeSccjr2pFocMAdefpS46YJ4zScEcjryP8ACnDOOD7n/wCvUMY4Y4/nTsgDHqCT259/emDJ4PB56c0/ggnPp+dIpDlIOOPzH+FTIOnpnHoKiXGScngD8qlUZwP14H51D1KJ04B5qVOp544Gf51EgwR34qZeMfgTk01sUSLnJ6cZp68Ec/5/CmAkY9Cc+ufWpBjJ7cfl25qkhjhgEDHvTs5AyM8f5FNBPIHOP8Kdgn/Pf1oSC44A+nXnqRmpFHQY9+xFRqSBx6fTmnjAIycd8Hv+FMCRck+3B4qZAMkY6496hXJJPbqO3+c1MgHJ/DH/AOqk0UixFgdz/X2q0MZBJzx+vvVePBAwMZ49wOtTgjIz6HP/ANaqQmSLgDOR1+lKCD7VEDjP5/5FOzjp071aIaJAwA9iT3z+dBbHv+PamA9eOuf/ANdNyOQeO2ev5U0SyXfgc46/h+NKGyTkcHHT9Kh3DgHjt9e4pd5AH8qaEP3cHnjn8aN4BHp0z6enFRFsd/f0z600sOvahsRNvP4dOKQtnPOOQfw9jURfng00vznI445oHYnEnHv06dKeHBwScZ/D8Kqh8k4I9PxpwYZB46f5yKTCxaDZwMn1xz+dPDc9PSqyuTjgZqQMMjn/AOvSeg0i1Gcnpj3/ABqdCSCeeOKqxkHJHpx/hVlD04Pr/WkmS0WF5/Ht/Op0HAOPUY9+5qBDkj86sKT1zz/nNWjOSJEHAz14/wAin4689f8AP501R0OOTipVH+eM1otTKQgAyPbA/ClIBJGSf504AZ9BzjtTgvPv/WrRmyMqOmMn/PWk2jHToOvSpsEZ45pCMkjH+H1qrENkDLjOOlRsgI6c9KtFSM8cflTNuT7dO9UkS2VShz045x/jTTFknjGOn+NWygOeOf5UmwcZAH+FUkIqGLPAH+fypjRgZ4x3+v8A9aruzPJGBnP4U0oM8gn0+lWhXKRi5PHUg/h0pPKxjAHt2q2Yx6f/AKqPKBJxx1piKoiwc4+vpS+VnHH9Pzq2IjkZAI/zmniIYHHPaqQmyoIiR0+vpn/GnCEEDjnj24q35RBPHXrThFg07ktlURYxx1pwiB6j+VW | 40,960 |
/HW15/dz15.ipynb | 61869f6d4c66f0556edf514fb933097df9e90c00 | [] | no_license | checheanya/HSE_bioinformatics | https://github.com/checheanya/HSE_bioinformatics | 0 | 0 | null | null | null | null | Jupyter Notebook | false | false | .py | 21,571 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# ### Problem 1
# +
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
from sklearn.pipeline import Pipeline
from sklearn.preprocessing import StandardScaler
from sklearn.neighbors import KNeighborsClassifier
from sklearn.model_selection import train_test_split
from sklearn.metrics import accuracy_score
# -
brca = pd.read_csv('BRCA_pam50.tsv', index_col = 0, sep = '\t')
brca.head(3)
brca['Subtype'].value_counts()
# дисбаланс
X = brca.iloc[:, :-1].to_numpy()
y = brca["Subtype"].to_numpy()
X_train, X_test, y_train, y_test = train_test_split(
X, y, stratify=y, test_size=0.2, random_state=17
)
# +
model = Pipeline([
("scaler", StandardScaler()),
("clf", KNeighborsClassifier(n_neighbors=1, weights="distance", p=2))
])
# p=1 = manhattan_distance (l1), and euclidean_distance (l2) for p=2
# как-то грустно и неправильно с 1 соседом делать......
model.fit(X_train, y_train)
y_pred = model.predict(X_test)
print(accuracy_score(y_pred, y_test))
# -
# ### Problem 2
for typ in np.unique(y_test).tolist():
print(typ,
sum(y_pred[np.where(y_test == typ)] == y_test[np.where(y_test == typ)]) / len(y_test[np.where(y_test == typ)])
)
# Поскольку мы с самого начала выявили четкий дисбаланс классов - люминал А сильно больше - эти результаты ожидаемы. На люминал А модель показала высокую точность и это внесло бОльший вклад в общую точность. На других же классах точность небольшая, но засчет их небольшого количества, их вклад в общую точность незначительный.
# ### Problem 3
# +
from sklearn.neighbors import NearestCentroid
model_centr = Pipeline([
("scaler", StandardScaler()),
("clf", NearestCentroid())
])
model_centr.fit(X_train, y_train)
y_pred_centr = model_centr.predict(X_test)
print(accuracy_score(y_pred_centr, y_test))
# -
for typ in np.unique(y_test).tolist():
print(typ,
sum(y_pred_centr[np.where(y_test == typ)] == y_test[np.where(y_test == typ)]) / len(y_test[np.where(y_test == typ)])
)
# Общая точность осталась примерно такая же, но точность по классам изменилась в лучшую сторону (хотя бы нет нулей...).
# ### Problem 4
# +
df = pd.DataFrame({'x': [1, 1, 2], 'y': [1, 2.5, 2], 'class': [0, 1, 2]})
sns.scatterplot(x = 'x', y = 'y', data = df, hue = 'class');
# +
print('L1', '\n', 'first',
df['x'][1] - df['x'][0] + df['y'][1] - df['y'][0], '\n', 'second',
df['x'][2] - df['x'][0] + df['y'][2] - df['y'][0]
)
print('L2', '\n', 'first',
np.sqrt((df['x'][1] - df['x'][0])**2 + (df['y'][1] - df['y'][0])**2), '\n', 'second',
np.sqrt((df['x'][2] - df['x'][0])**2 + (df['y'][2] - df['y'][0])**2)
)
| 2,914 |
/Lim-m3.ipynb | 0cf6fd8cda1fa73241efe88f1ee27d7caac77b37 | [] | no_license | evlim/Optimization | https://github.com/evlim/Optimization | 0 | 0 | null | null | null | null | Jupyter Notebook | false | false | .py | 581,100 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Excercise 3.32
# Evan Lim
# ## Part A
# 
# ### Model Setup
# +
from gurobipy import *
m = Model('ex332')
OakReq = [17,0,5,0] # bd. ft. of oak
PineReq = [0,30,0,13] # bd. ft. of oak
Prices = [1000,1000,300,300] # prices per unit
OakMax = 25000
PineMax = 20000
# -
# ### Decision Variables
N = 4 #
Xnames = ['Oak Tables','Pine Tables','Oak Chairs','Pine Chairs']
X = []
for i in range(N):
X.append(m.addVar(vtype = GRB.CONTINUOUS, lb = 0.0, name = Xnames[i]))
m.update()
# ### Constraints
# $\sum_{j=0}^N{x_j c_j}$
m.addConstr(quicksum(X[j]*OakReq[j] for j in range(N)), GRB.LESS_EQUAL, OakMax, "Oak")
m.addConstr(quicksum(X[j]*PineReq[j] for j in range(N)), GRB.LESS_EQUAL, PineMax, "Pine")
m.update()
# ### Objective
m.setObjective(quicksum(X[j]*Prices[j] for j in range(N)), GRB.MAXIMIZE)
m.update()
# ### Model Output
m.optimize()
print(f"The maximized revenue is ${m.objVal:,.0f}\n")
print("The optimal production schedule is: ")
for i in range(N):
print(f"{X[i].varName}\t: {X[i].x:.0f}")
# ## Part b
k = [i/10+1 for i in range(-3,4)]
k
m.setParam('OutputFlag', False) # hide Gurobi console output
rev = {} # optimal revenue
for pine in k:
for oak in k:
p = [Prices[0]*oak, Prices[1]*pine, Prices[2]*oak, Prices[3]*pine]
m.setObjective(quicksum(X[j]*p[j] for j in range(N)), GRB.MAXIMIZE)
m.update()
m.optimize()
rev[(pine,oak)] = m.objVal
import pandas as pd
pd.set_option('display.float_format', lambda x: '%.0f' % x)
# +
d = {}
for pine in k:
d[f"Pine {pine}"] = [rev[pine,oak] for oak in k]
df = pd.DataFrame.from_dict(d)
df.index = [f"Oak {oak}" for oak in k]
df
# -
import matplotlib.pyplot as plt
import numpy as np
import warnings
# %matplotlib inline
with warnings.catch_warnings():
warnings.simplefilter("ignore")
df.plot()
# ##### Remarks
# As pine and oak products are varied from -30% to +30%, the optimal (maximized) revenue changes linearly.
#
>>Param :: D0 := 10.d0 <<
# >>Param :: D1 := 0.3d0 <<
# >>Param :: oneHalf := 0.5d0 <<
# ```
# Of course, numerical prefactors can often be absorbed in physical parameters. (An alternative to the declaration above would be to define a renormalized $D_1^* = D_1/2$.) However, this is not always possible, which is why we illustrate how to create a named parameter for numerical prefactors.
#
# ## List of variables
# ### Overview
# Coral require that the user defines four kind of variables:
# + `>>linear_variable_kxky` are fluctuating components of fields, that are linearly coupledIt is natural to think of these components in spectral space.
# + `>>linear_variable_mean` are horizontal-mean components of fields (i.e. functions of the vertical coordinate `z` only), that are linearly coupled. As with `>>linear_variable_mean`, those quantities are intrinsically spectral
# + `>>linear_variable_full` corresponds to fields that are computed in physical space. They are linear combinations of fluctuating and / or horizontal-mean components. (That is to say that they are necessarily formed by combining members from `>>linear_variable_kxky` and `>>linear_variable_mean`.) Their reason of being is to be building blocks for quadratic variables (also in physical space, see next bullet). Since they are computed in physical space, these `full` variables are available for output (in the forms of volumes, slices, profiles, or timeseries).
# + `>>quadratic_variable` are products of `>>linear_variable_full` and may represent advection. These `quadratic` variables are computed in physical space, and therefore are also available for output (in the forms of volumes, slices, profiles, or timeseries).
#
# ### Illustration on the diffusion equation example
#
#
# We introduce the horizontal-average $\overline{\phi}(z)=\left\langle \phi\right\rangle_{x,y}$ and the fluctuations around this mean $\widetilde{\phi}(x,y,z)=\phi(x,y,z) - \overline{\phi}(z)$, which are treated as distinct linear variables. These variables, which we denote `phiMean` and `phi`, obey slightly different equations (obtained by projecting the diffusion equation above):
# \begin{gather}
# \frac{\partial}{\partial t}\iint \widetilde{\phi}= D_0 \widetilde{\phi} + \iint D_0 \nabla^2_\perp \widetilde{\phi} + \frac{D_1}{2} \iint \nabla_\perp^2 \widetilde{\phi^2}+ \frac{D_1}{2} \widetilde{\phi^2} \nonumber \\
# \frac{\partial}{\partial t}\iint \overline{\phi}= D_0 \overline{\phi} + \frac{D_1}{2} \overline{\phi^2} \nonumber
# \end{gather}
# Of course, the full field $\phi$, denoted `phiFull`, is recovered by merely computing the sum `phiMean + phi`. Finally, the quadratic variable `phi2` represents the square of `phiFull`. Hence the declaration of variables:
# ```
# >>====================================<<
# >>linear_variable_kxky :: phi <<
# >>linear_variable_mean :: phiMean <<
# >>====================================<<
# >>linear_variable_full :: phiFull <<
# >>linear_variable_full_build :: + phiMean <<
# >>linear_variable_full_build :: + phi <<
# >>====================================<<
# >>quadratic_variable :: phi2 := phiFull.phiFull <<
# >>====================================<<
# ```
# ## Set of equations
# ### The finite wavenumber system
# We start with the finite-wavenumber governing equation
# \begin{equation}
# \frac{\partial}{\partial t}\iint \widetilde{\phi}= D_0 \widetilde{\phi} + \iint D_0 \nabla^2_\perp \widetilde{\phi} + \frac{D_1}{2} \iint \nabla_\perp^2 \widetilde{\phi^2}+ \frac{D_1}{2} \widetilde{\phi^2} \nonumber \end{equation}
# This equation is second order in z (indicated below by `>>new_equation :: 2 <<`). Linearly, only $\mathtt{phi}=\widetilde{\phi}$ is involved. This variable need to be supplemented with two boundary conditions, for instance a no-flux top and bottom: $\partial_z \phi = 0$ at $z=0,1$ (encoded by integer `21` in Coral).
#
# ```
# >>====================================<<
# >>add_set_of_coupled_kxky_equations <<
# >>linearly_coupled_var :: phi <<
# >>add_BC_for_this_var :: 21 <<
# >>new_equation :: 2 <<
# >>add_d/dt_term :: + Iz.Iz.phi <<
# >>add_rhs_linear :: + D0.phi <<
# >>add_rhs_linear :: + D0.Iz.Iz.dx.dx.phi <<
# >>add_rhs_linear :: + D0.Iz.Iz.dy.dy.phi <<
# >>add_rhs_NL :: + D1.oneHalf.Iz.Iz.dx.dx.phi2 <<
# >>add_rhs_NL :: + D1.oneHalf.Iz.Iz.dy.dy.phi2 <<
# >>add_rhs_NL :: + D1.oneHalf.phi2 <<
# >>====================================<<
# ```
# ### The finite wavenumber system
# Next, we enter the horizontally-averaged equation:
# \begin{equation}
# \frac{\partial}{\partial t}\iint \overline{\phi}= D_0 \overline{\phi} + \frac{D_1}{2} \overline{\phi^2} \nonumber \end{equation}
# This equation is also second order in z. Linearly, only $\mathtt{phiMean}=\overline{\phi}$ is involved, and for consistency this variable follows the same stress-free boundary condition.
#
# ```
# >>====================================<<
# >>add_set_of_coupled_kxky_equations <<
# >>linearly_coupled_var :: phiMean <<
# >>add_BC_for_this_var :: 21 <<
# >>new_equation :: 2 <<
# >>add_d/dt_term :: + Iz.Iz.phiMean <<
# >>add_rhs_linear :: + D0.phiMean <<
# >>add_rhs_NL :: + D1.oneHalf.phi2 <<
# >>====================================<<
# ```
# ### End of file
# Once the sets of equations have been define, we must indicate that we have reached the end of file by adding the line:
# ```
# >>EOF<<
# ```
#
# ## Closing remarks
#
# In this notebook, we have learned the basics of implementing equations in Coral:
# + defining parameters
# + distinguishing between linearly terms and quadratic contributions (and how to build the latter)
# + distinguishing between horizontally-averaged components and fluctuations around this mean
#
# However, we worked on a simple example that consisted in a unique equation for a unique variable. In the other notebooks, we ramp up the complexity of the equations by introducing coupled sets of equations (e.g. Navier-Stokes equations).
| 8,230 |
/Lab Assignment GridWorld - Python code.ipynb | b475e9a4bd0d64ae92e7676a35ec3926c1cd56df | [] | no_license | maxtaylordavies/COMP70028 | https://github.com/maxtaylordavies/COMP70028 | 0 | 0 | null | null | null | null | Jupyter Notebook | false | false | .py | 18,631 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# + [markdown] id="dvjrFo0_0IXN" colab_type="text"
# #IMPORTING LIBRARIES
# + id="KPgPHSveyeNr" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 52} outputId="edf3d44c-ec93-444f-83de-02bd961657a2"
import keras
keras.__version__
# + id="rU527ns2Xn7Y" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 51} outputId="8ccd69e8-0338-4d81-a9e7-692024293824"
from keras.datasets import boston_housing
(train_data, train_targets), (test_data, test_targets) = boston_housing.load_data()
# + id="EaKNObWTely2" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 34} outputId="453106da-e0ea-4aef-dfa4-8592c4753d9f"
train_data.shape
# + id="hYIEcnhYerYJ" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 34} outputId="d479f3e3-5ad1-4926-b439-53bfdc0272ec"
test_data.shape
# + id="KZ2gKQzvew4P" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 646} outputId="a724e754-2bb6-4e03-dc39-858d0933c85c"
train_targets
# + id="Sld525vFfARN" colab_type="code" colab={}
mean = train_data.mean(axis=0)
train_data -= mean
std = train_data.std(axis=0)
train_data /= std
test_data -= mean
test_data /= std
# + id="RitxSdZSfEcW" colab_type="code" colab={}
from keras import models
from keras import layers
def build_model():
# Because we will need to instantiate
# the same model multiple times,
# we use a function to construct it.
model = models.Sequential()
model.add(layers.Dense(64, activation='relu',
input_shape=(train_data.shape[1],)))
model.add(layers.Dense(64, activation='relu'))
model.add(layers.Dense(1))
model.compile(optimizer='rmsprop', loss='mse', metrics=['mae'])
return model
# + id="ocu0J1bpyeNx" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 85} outputId="efdb2b3a-a6ec-4041-cc56-602a9f86923a"
import numpy as np
k = 4
num_val_samples = len(train_data) // k
num_epochs = 100
all_scores = []
for i in range(k):
print('processing fold #', i)
# Prepare the validation data: data from partition # k
val_data = train_data[i * num_val_samples: (i + 1) * num_val_samples]
val_targets = train_targets[i * num_val_samples: (i + 1) * num_val_samples]
# Prepare the training data: data from all other partitions
partial_train_data = np.concatenate(
[train_data[:i * num_val_samples],
train_data[(i + 1) * num_val_samples:]],
axis=0)
partial_train_targets = np.concatenate(
[train_targets[:i * num_val_samples],
train_targets[(i + 1) * num_val_samples:]],
axis=0)
# Build the Keras model (already compiled)
model = build_model()
# Train the model (in silent mode, verbose=0)
model.fit(partial_train_data, partial_train_targets,
epochs=num_epochs, batch_size=1, verbose=0)
# Evaluate the model on the validation data
val_mse, val_mae = model.evaluate(val_data, val_targets, verbose=0)
all_scores.append(val_mae)
# + id="qKIKZ5LhOusF" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 34} outputId="e48a344f-8253-4167-efe6-b885537e059e"
all_scores
# + id="cMKuaUa5O0dF" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 34} outputId="5f3b666f-c12b-4777-df64-33e0ab21af2e"
np.mean(all_scores)
# + id="771zNhPsPyl_" colab_type="code" colab={}
from keras import backend as K
# Some memory clean-up
K.clear_session()
# + id="DpRB1IbOP2SQ" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 85} outputId="5ac4609a-11c1-445a-9fff-c595e7914e4e"
num_epochs = 500
all_mae_histories = []
for i in range(k):
print('processing fold #', i)
# Prepare the validation data: data from partition # k
val_data = train_data[i * num_val_samples: (i + 1) * num_val_samples]
val_targets = train_targets[i * num_val_samples: (i + 1) * num_val_samples]
# Prepare the training data: data from all other partitions
partial_train_data = np.concatenate(
[train_data[:i * num_val_samples],
train_data[(i + 1) * num_val_samples:]],
axis=0)
partial_train_targets = np.concatenate(
[train_targets[:i * num_val_samples],
train_targets[(i + 1) * num_val_samples:]],
axis=0)
# Build the Keras model (already compiled)
model = build_model()
# Train the model (in silent mode, verbose=0)
history = model.fit(partial_train_data, partial_train_targets,
validation_data=(val_data, val_targets),
epochs=num_epochs, batch_size=1, verbose=0)
mae_history = history.history['mae']
all_mae_histories.append(mae_history)
# + id="AecAE5zGP5lT" colab_type="code" colab={}
average_mae_history = [
np.mean([x[i] for x in all_mae_histories]) for i in range(num_epochs)]
# + id="07LmXrpVP8lD" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 279} outputId="89d98770-fe2c-4546-cd8d-0fa6756e4089"
import matplotlib.pyplot as plt
plt.plot(range(1, len(average_mae_history) + 1), average_mae_history)
plt.xlabel('Epochs')
plt.ylabel('Validation MAE')
plt.show()
# + id="DsHyDGsOOuwZ" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 279} outputId="53def0dd-a571-4c4f-fac0-d37a92aff932"
def smooth_curve(points, factor=0.9):
smoothed_points = []
for point in points:
if smoothed_points:
previous = smoothed_points[-1]
smoothed_points.append(previous * factor + point * (1 - factor))
else:
smoothed_points.append(point)
return smoothed_points
smooth_mae_history = smooth_curve(average_mae_history[10:])
plt.plot(range(1, len(smooth_mae_history) + 1), smooth_mae_history)
plt.xlabel('Epochs')
plt.ylabel('Validation MAE')
plt.show()
# + id="7wnyknfsOyXb" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 34} outputId="20c3fe80-55d7-40a7-f7da-56af121639cb"
model = build_model()
# Train it on the entirety of the data.
model.fit(train_data, train_targets,
epochs=80, batch_size=16, verbose=0)
test_mse_score, test_mae_score = model.evaluate(test_data, test_targets)
# + id="2pzFbTKhO0q0" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 34} outputId="e219b197-c1c3-424f-943e-fc3b8eddee7f"
test_mae_score
nal.feature_importances_), data.columns),
reverse=True))
# ### Task 5: Final summary
# Write a super-brief (under 200 words) summary of your process, the dataset and your model (including what features were most important).
#
# This is an investigation on lethalities of heart failure. First, we inspected the data and concluded the data is in good quality. We then explored the data to grasp basic information: we know there are 13 features (6 qualitative and 7 quantitative), ~300 rows). We also noticed the data is in class imbalance, as number of non-deaths are 2 times more than deaths. Next, we explored further in hopes to find hypotheses on dominant predictors of death, with visualisations in between, ending with a heatmap to show correlation between all variables. Then, to do the modelling, we first split the data in 80-20, and used Naive Bayes as a comparison towards the Random Forests we will focus on. We focused on grid-searching several traditionally dominant hyperparameters (eg. n_estimators), and randomized the search to achieve a lower computational power while not harming too much on the performance. At last, while evaluating the prediction, we used ROC_AUC score since the data is intrinsically in imbalance. Note that the feature importance ranking (as part of output of final model) claims time, serum_creatinine and ejection_fraction as 3 most important factors, which coincides with the final row of the heatmap.
#And append it to the valid state locations if it is a valid state (ie not absorbing)
if(self.is_location(loc)):
locs.append(loc)
# Get an array with the neighbours of each state, in terms of locations
local_neighbours = [self.get_neighbour(loc,direction) for direction in ['nr','ea','so', 'we']]
neighbour_locs.append(local_neighbours)
# translate neighbour lists from locations to states
num_states = len(locs)
state_neighbours = np.zeros((num_states,4))
for state in range(num_states):
for direction in range(4):
# Find neighbour location
nloc = neighbour_locs[state][direction]
# Turn location into a state number
nstate = self.loc_to_state(nloc,locs)
# Insert into neighbour matrix
state_neighbours[state,direction] = nstate;
# Translate absorbing locations into absorbing state indices
absorbing = np.zeros((1,num_states))
for a in self.absorbing_locs:
absorbing_state = self.loc_to_state(a,locs)
absorbing[0,absorbing_state] =1
return locs, state_neighbours, absorbing
def loc_to_state(self,loc,locs):
#takes list of locations and gives index corresponding to input loc
return locs.index(tuple(loc))
def is_location(self, loc):
# It is a valid location if it is in grid and not obstacle
if(loc[0]<0 or loc[1]<0 or loc[0]>self.shape[0]-1 or loc[1]>self.shape[1]-1):
return False
elif(loc in self.obstacle_locs):
return False
else:
return True
def get_neighbour(self,loc,direction):
#Find the valid neighbours (ie that are in the grif and not obstacle)
i = loc[0]
j = loc[1]
nr = (i-1,j)
ea = (i,j+1)
so = (i+1,j)
we = (i,j-1)
# If the neighbour is a valid location, accept it, otherwise, stay put
if(direction == 'nr' and self.is_location(nr)):
return nr
elif(direction == 'ea' and self.is_location(ea)):
return ea
elif(direction == 'so' and self.is_location(so)):
return so
elif(direction == 'we' and self.is_location(we)):
return we
else:
#default is to return to the same location
return loc
###########################################
# + button=false deletable=true new_sheet=false run_control={"read_only": false}
grid = GridWorld()
### Question 1 : Change the policy here:
Policy= np.zeros((grid.state_size, grid.action_size))
print("The Policy is : {}".format(Policy))
val = 0 #Change here!
print("The value of that policy is : {}".format(val))
# + button=false deletable=true new_sheet=false run_control={"read_only": false}
# Using draw_deterministic_policy to illustrate some arbitracy policy.
Policy2 = np.zeros(22).astype(int)
Policy2[2] = 3
Policy2[6] = 2
Policy2[18] = 1
grid.draw_deterministic_policy(Policy2)
# + button=false deletable=true new_sheet=false run_control={"read_only": false}
# + button=false deletable=true new_sheet=false run_control={"read_only": false}
# + button=false deletable=true new_sheet=false run_control={"read_only": false}
| 11,730 |
/notebooks/07_PyTorch.ipynb | c5f7a0e4baf902a45812de5d4eb22ea40918418e | [
"MIT"
] | permissive | Mike4cozy/practicalAI | https://github.com/Mike4cozy/practicalAI | 0 | 0 | MIT | 2019-03-22T08:11:38 | 2019-03-22T08:11:34 | null | Jupyter Notebook | false | false | .py | 19,830 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Notes on Voit et al 2019 A Black-Hole Feedback Valve in Massive Galaxies
#
# ## Notes from Abstract:
#
# AGN release enough energy to inhibit cooling of hot gas, preventing it from condensing into stars. But energy arguments alone do not account for the effectiveness of quenching in high-mass galaxies. Optical observations show quenching is more closely related to a galaxy's central stellar velocity dispersion than anything else.
#
# This paper shows that high central velocity dispersion maximizes the efficacy of AGN feedback.
#
# To remain quenched, a galaxy must continually sweep out the gas ejected from its aging stars. Supernova heating can do this, as long as the AGN reduces the gas pressure of the CGM.
#
# CGM pressure acts as the control knob on a valve that regulates AGN feedback and the feedback power self-adjusts so that it suffices to lift the CGM out of the galaxy's potential well. Supernova heating then drives a galactic outflow that remains homogenous for large enough velocity dispersion.
#
# Feedback in galaxies with a lower velocity dispersion tends to result in convective circulation and accumulation of multiphase gas within the galaxy.
# ## Notes from the Introduction:
#
# Three different gas sources must be disrupted in order to prevent star formation from starting up again.
# 1) Cold streams: cold gas coming into the galaxy from dark matter filaments.
# - Models of galaxy evolution contend that quiescent central galaxies have hot halos that disrupt cold streams and that quiescent central galaxies orbiting the central one cannot access cold streams due to their displacement from the center.
#
# 2) Cooling flows: Radiative cooling of dense gas inside the halo supplies the galaxy.
# - Models contend that accreting cold gas onto the central SMBH releases enough energy to offset radiative losses.
#
# 3) Stellar Mass loss: Dying stars shed surplus gas.
# - Supernova heating can push this gas outside the galaxy, but CGM pressure limits the ability to do so. CGM confining pressure determines outflow gas density and radiative losses.
#
# ## Questions:
# What is a "cooling catastrophe"?
#
#
# # Isothermal Hydrostatic Galaxy Model
#
# Approximation is valid for galaxies with a velocity field that is sufficiently subsonic.
#
d_index()`.
# + id="9Rb9FF-hkgh6" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 9908} outputId="0544f264-e921-4feb-a19d-1a669256be22"
X_train[7]
# + id="qtz01B01dtot" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 107} outputId="5090d07a-0bb1-4c03-e687-eac959155e81"
# Map word IDs back to words
word2id = imdb.get_word_index()
id2word = {i: word for word, i in word2id.items()}
print("--- Review (with words) ---")
print([id2word.get(i, " ") for i in X_train[7]])
print("--- Label ---")
print(y_train[7])
# + [markdown] id="kNL0UO5adtoy" colab_type="text"
# Unlike our Bag-of-Words approach, where we simply summarized the counts of each word in a document, this representation essentially retains the entire sequence of words (minus punctuation, stopwords, etc.). This is critical for RNNs to function. But it also means that now the features can be of different lengths!
#
# #### Question: Variable length reviews
#
# What is the maximum review length (in terms of number of words) in the training set? What is the minimum?
#
# #### Answer:
#
# ...
#
#
# ### TODO: Pad sequences
#
# In order to feed this data into your RNN, all input documents must have the same length. Let's limit the maximum review length to `max_words` by truncating longer reviews and padding shorter reviews with a null value (0). You can accomplish this easily using the [`pad_sequences()`](https://keras.io/preprocessing/sequence/#pad_sequences) function in Keras. For now, set `max_words` to 500.
# + id="eYdFncirdto0" colab_type="code" colab={}
from keras.preprocessing import sequence
# Set the maximum number of words per document (for both training and testing)
max_words = 500
# TODO: Pad sequences in X_train and X_test
X_train_p = sequence.pad_sequences(X_train, maxlen=max_words)
X_test_p = sequence.pad_sequences(X_test, maxlen=max_words)
# + id="umfaKW03mbsL" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 34} outputId="8f75df5e-ea3d-47c2-f0db-7b3b1661d7c7"
len(X_train[7])
# + id="zvJk8CXLnq-f" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 34} outputId="9c783c82-2cd7-41e7-c4b8-8dd1ca4898c2"
X_train[7][-10:]
# + id="mqKkJv50neYn" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 52} outputId="04b519b8-8859-4c9a-c7e0-f79c6e875de7"
X_train_p[7][-10:]
# + [markdown] id="ZjR3OkmCdto6" colab_type="text"
# ### TODO: Design an RNN model for sentiment analysis
#
# Build your model architecture in the code cell below. We have imported some layers from Keras that you might need but feel free to use any other layers / transformations you like.
#
# Remember that your input is a sequence of words (technically, integer word IDs) of maximum length = `max_words`, and your output is a binary sentiment label (0 or 1).
# + id="xuVu98codto8" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 263} outputId="6c7209f0-d22f-41f1-8fd6-6917fca102a3"
from keras.models import Sequential
from keras.layers import Embedding, LSTM, Dense, Dropout
# TODO: Design your model
embedding_size = 32
model = Sequential()
model.add(Embedding(vocabulary_size, embedding_size, input_length=max_words))
model.add(LSTM(100))
model.add(Dense(1, activation='sigmoid'))
print(model.summary())
# + [markdown] id="aUMPik8JdtpD" colab_type="text"
# #### Question: Architecture and parameters
#
# Briefly describe your neural net architecture. How many model parameters does it have that need to be trained?
#
# #### Answer:
#
# ...
#
# ### TODO: Train and evaluate your model
#
# Now you are ready to train your model. In Keras world, you first need to _compile_ your model by specifying the loss function and optimizer you want to use while training, as well as any evaluation metrics you'd like to measure. Specify the approprate parameters, including at least one metric `'accuracy'`.
# + id="Z9BsHerNdtpG" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 160} outputId="d58e2c9b-458a-4b46-c3ac-c146b702b2a9"
# TODO: Compile your model, specifying a loss function, optimizer, and metrics
model.compile(loss='binary_crossentropy', optimizer='adam', metrics=['accuracy'])
# + [markdown] id="pf0bWfQBdtpL" colab_type="text"
# Once compiled, you can kick off the training process. There are two important training parameters that you have to specify - **batch size** and **number of training epochs**, which together with your model architecture determine the total training time.
#
# Training may take a while, so grab a cup of coffee, or better, go for a hike! If possible, consider using a GPU, as a single training run can take several hours on a CPU.
#
# > **Tip**: You can split off a small portion of the training set to be used for validation during training. This will help monitor the training process and identify potential overfitting. You can supply a validation set to `model.fit()` using its `validation_data` parameter, or just specify `validation_split` - a fraction of the training data for Keras to set aside for this purpose (typically 5-10%). Validation metrics are evaluated once at the end of each epoch.
# + id="yqSPTlxTdtpO" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 213} outputId="e9b457d6-32a4-4a99-a738-cd5f4ccc43cc"
# TODO: Specify training parameters: batch size and number of epochs
batch_size = 64
num_epochs = 3
# TODO(optional): Reserve/specify some training data for validation (not to be used for training)
X_valid, y_valid = X_train_p[:batch_size], y_train[:batch_size]
X_train2, y_train2 = X_train_p[batch_size:], y_train[batch_size:]
# TODO: Train your model
model.fit(X_train2, y_train2,
validation_data=(X_valid, y_valid),
batch_size=batch_size,
epochs=num_epochs)
# + id="bGQ1sqordtpV" colab_type="code" colab={}
import os
# Save your model, so that you can quickly load it in future (and perhaps resume training)
model_file = "rnn_model.h5" # HDF5 file
model.save(os.path.join('/content', model_file))
# Later you can load it using keras.models.load_model()
#from keras.models import load_model
#model = load_model(os.path.join(cache_dir, model_file))
# + [markdown] id="jmD6IqVBdtpd" colab_type="text"
# Once you have trained your model, it's time to see how well it performs on unseen test data.
# + id="ssYWxFv7dtph" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 34} outputId="90771697-bdb2-4b78-9f56-94b330adbc75"
# Evaluate your model on the test set
scores = model.evaluate(X_test_p, y_test, verbose=0) # returns loss and other metrics specified in model.compile()
print("Test accuracy:", scores[1]) # scores[1] should correspond to accuracy if you passed in metrics=['accuracy']
# + [markdown] id="p848CCCcdtpr" colab_type="text"
# #### Question: Comparing RNNs and Traditional Methods
#
# How well does your RNN model perform compared to the BoW + Gradient-Boosted Decision Trees?
#
# #### Answer:
#
# ...
#
# ## Extensions
#
# There are several ways in which you can build upon this notebook. Each comes with its set of challenges, but can be a rewarding experience.
#
# - The first thing is to try and improve the accuracy of your model by experimenting with different architectures, layers and parameters. How good can you get without taking prohibitively long to train? How do you prevent overfitting?
#
# - Then, you may want to deploy your model as a mobile app or web service. What do you need to do in order to package your model for such deployment? How would you accept a new review, convert it into a form suitable for your model, and perform the actual prediction? (Note that the same environment you used during training may not be available.)
#
# - One simplification we made in this notebook is to limit the task to binary classification. The dataset actually includes a more fine-grained review rating that is indicated in each review's filename (which is of the form `<[id]_[rating].txt>` where `[id]` is a unique identifier and `[rating]` is on a scale of 1-10; note that neutral reviews > 4 or < 7 have been excluded). How would you modify the notebook to perform regression on the review ratings? In what situations is regression more useful than classification, and vice-versa?
#
# Whatever direction you take, make sure to share your results and learnings with your peers, through blogs, discussions and participating in online competitions. This is also a great way to become more visible to potential employers!
# + id="eEdQjXgVdtpt" colab_type="code" colab={}
| 11,174 |
/kaggle-teams/kaggle_teams_preso.ipynb | 99f10a5f6678eabe5c623b51d62aaed740bcf792 | [
"BSD-2-Clause"
] | permissive | Anhmike/kaggle | https://github.com/Anhmike/kaggle | 0 | 0 | null | null | null | null | Jupyter Notebook | false | false | .py | 13,038 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# + [markdown] slideshow={"slide_type": "slide"}
# # Kaggle Teams !!!
# [](https://kaggle.com "Kaggle" target="_blank")
# ## A quantitative and qualitative look at Kaggle team performance: 2012-2016
# ## How they form, perform, and work
# + [markdown] slideshow={"slide_type": "slide"}
# # What's the goal?
#
# - Strategizing to become a Master?
# - Is it better to join a team or go it alone?
# - Wondering who, where, and what goes in to a winning team?
# - Wondering how to get invited to a team?
# - Wondering how to create a team?
# - What are the best tools, stacks?
# - How to achieve top results?
# + [markdown] slideshow={"slide_type": "slide"}
# # Me - [paulperry](https://www.kaggle.com/paulperry)
#
# [https://www.kaggle.com/paulperry](https://www.kaggle.com/paulperry)
#
# [](https://www.kaggle.com/paulperry "paulperry" target="_blank")
#
#
# + [markdown] slideshow={"slide_type": "slide"}
# # Strategizing to become a Master
# - **Don't chase the leaderboard**: You are at risk of overfiting.
# - **Don't chase rank** Don't be a lazy kaggler and chase rank by submitting other people's scripts.
# - **Read the Forums**: seems obvious but every little nugget counts.
# - **Share some scripts**: scripts do provide great opportunities for code sharing and learning, but beat those scripts
# - **[How long does it take to place first?](https://www.kaggle.com/shujian/how-long-it-takes-to-win-1st-on-kaggle)** About a year.
#
# + [markdown] slideshow={"slide_type": "subslide"}
# # [Scripty McScriptface the Lazy Kaggler](https://www.kaggle.com/dvasyukova/scripty-mcscriptface-the-lazy-kaggler)
#
# [](https://www.kaggle.com/dvasyukova/scripty-medals "" target="_blank")
# + [markdown] slideshow={"slide_type": "slide"}
# # Are you better off solo?
#
# **[solo wins are rare](https://www.kaggle.com/reaces/are-you-better-of-solo/output "solo wins")**, and only after a lot of submissions
#
# [](https://www.kaggle.com/rachelsunny/do-teams-perform-better-than-soloists)
# + [markdown] slideshow={"slide_type": "slide"}
# # Kagglers are teaming up more
# [](https://www.kaggle.com/jeongyoonlee/kaggle-competitions-over-time-w-updated-data/notebook "Number of Teams Competiting in Kaggle Competitions")
#
# * Credit [Jeong-Yoon Lee](https://www.kaggle.com/jeongyoonlee)
#
#
# + [markdown] slideshow={"slide_type": "slide"}
# # And teams are winning more
# - In [May 13, 2015](http://blog.kaggle.com/2015/05/13/improved-kaggle-rankings/ "" target="_blank"), Kaggle updated the ranking system to penalize teams less than before.
# - Recent competitions (after May 15, 2015) feature big and rich data sets conducive to blending has meant increased team-ups in the top of the leaderboard.
# - [TL;DR: New masters are still being made](https://www.kaggle.com/willieliao/making-the-master-tier-top-10-version?scriptVersionId=399231). Keep learning, climb the leaderboard, and team up.
#
# [](https://www.kaggle.com/willieliao/making-the-master-tier-top-10-version?scriptVersionId=399231 "" target="_blank")
# + [markdown] slideshow={"slide_type": "slide"}
# # When do you team up?
#
# - There is a lot to learn from the top data science practitioners.
# - They have dedicated a lot of time to develop their workflows,
# - have great intuition of what methods are likely to work,
# - and are efficient in their use of time.
# - “strategic” teaming: somewhere after 1/3 of the competition.
# - You plan to work together, learn, cooperate.
# - You may select teammates based on complimentary strengths.
# - You also can see if potential teammate actively participates in the competition and will not be a ballast.
# - Also by then you know if you want to dedicate time to competition yourself.
# - “tactical” teaming: Just before teaming deadline
# - Not much time to cooperate, but lets blend our models.
# - Makes sense for people close to each other on leaderboard especially near some threshold (teams 4 and 5 hoping to jump to second place after teaming-up) or near gold medal threshold
#
# 2. “Grandmaster problem” - needs individual gold medal.
# - Easy to do during recruiting competitions when teams are not allowed and scripts are limited.
#
# + [markdown] slideshow={"slide_type": "slide"}
# # Who do you team up with?
#
# - user ranking in current competition
# - user ranking in previous competition
# - someone with a different solution than yours:
# > "I usually ask the potential member about the solution and I try to figure out if its relatively different from my current solution. One thing that makes you win is diversity of solutions. Remember that blending 2 solution with low correlation is much better that blend 0.999 correlated solutions."
# [Titericz](https://www.kaggle.com/titericz) (aka giba1) [ref](https://docs.google.com/spreadsheets/d/19CBOB0a2HaGORDOBep0F8sltWr_Vv-h8Z0zAayqJGcY/edit#gid=95630921)
# - availability for the competition
# - user computer resources
# - someone who is active in the forums and who shares kernels
# - Someone with some experience, good work ethic, and positive attitude
# > "There has been exceptions, but in general would like someone that has played at least a few kaggle competitions . This is mainly to ensure that he/she understands the rules well, will not attempt cheating or do something funny by mistake .
# > I like a good work ethic. Good communications with positive attitude. There is no other requirement. In general I don't see a collaboration as a business thing. I am much more happy if after a potential merge we can be friends and long-lasting teammates with the group." [Kazanova](https://www.kaggle.com/kazanova)
# + [markdown] slideshow={"slide_type": "slide"}
# # How do you find team members?
# - [Top 100 Users with Most Team Memberships](https://www.kaggle.com/mlandry/top-100-users-with-most-team-memberships/code)
# - [Common Colloaborators](https://www.kaggle.com/mlandry/in-progress/code)
# - [Who are the most connected Kagglers?](https://www.kaggle.com/lbronchal/who-are-the-most-connected-kagglers)
# + [markdown] slideshow={"slide_type": "slide"}
# # The Kaggle Social Graph of Top Players
# 
# + [markdown] slideshow={"slide_type": "slide"}
# # How does the team organize their work?
#
# Kaggle team interview questions: What works? What doesn't work?
#
# 1. How do you select who you want to team with?
# - How do you communicate? Email, chat, video chat?
# - How do you share data (or features)?
# - Do you share code? If so, how?
# - Do you share code frameworks?
# - How do you decide to divide the work?
# - How do you avoid duplicative work? Or repeated work?
# - How do you avoid team members finding the same stuff?
# - How do you keep track of model performance?
# - How do you decide who gets to submit what on each day?
# - Who picks and and how do you pick the final submissions?
# - If you wanted to learn what worked on other Kaggle teams, what questions would you ask?
#
# + [markdown] slideshow={"slide_type": "slide"}
# # Interview with Lucas Eustaquio da Silva (Leustagos) 2/29/2016
#
# 1. How do you communicate? Email, chat, video chat? - **chat**
# - How do you share code? - **usually we don't, but when we do, its on dropbox**
# - How do you share data (or features)? - **csv on dropbox**
# - Do you share code frameworks? - **people don't like much to mess with another code**
# - How do you decide to divide the work? - **chat, but we usually have some slightly different approaches. its rare but we can suggest each other based on availability which ideas we can pursue first. Usually telling what i will do prevent others from doing the same**
# - How do you avoid duplicative work? Or repeated work? - **we don't avoid, but its not a big issue. i don't team up from the start so we can have distinct approaches when merging teams**
# - How do you avoid team members finding the same stuff? - **we don't**
# - How do you keep track of model performance? - **each one is responsible for keeping its versioning. i use git, some just duplicate and enumerate files. on each submission we describe which models we used to generate it**
# - How do you decide who gets to submit what on each day? - **common sense. we divide eauqly the number of submissions, but if someones need more he asks**
# - Who picks and and how do you pick the final submissions? - **the leader picks it. its a consensus. i never had any trouble to do it. with the right reasons its very easy to choose. Of course some times i don't agree and we just go with the majority. just don't be very picky and it will go smoothly. its very rare for me to not pick my best submission. except on some competitions that are too random.**
#
# Lucas
#
# + [markdown] slideshow={"slide_type": "slide"}
# # A proposed hierarchy of Kaggle team cooperation
#
# First, forget ["team camarderie"](http://analyticscosm.com/how-to-form-great-teams-of-data-scientists/). This is your last priority.
#
# 1. Agree on how and when to communicate: Slack, Discord, etc
# - Share ideas
# - Keep track of model performance across the team
# - Share resultsets
# - Agree on a CV strategy (fix fold indexes)
# - Ensemble model results: Usually one team member I responsible to keep the stack ensembling working.
# - Have a way to compare models
# - Share derived features
# - Share code fragments
# + [markdown] slideshow={"slide_type": "slide"}
# # DO: What teams could do better
#
# 1. Share code frameworks
# - Share cloud infrastructure
# - Share workload (split feature generation)
# - Review each other's work. Point to any obvious errors or omissions.
# - Share a computing instance
# - Share code platform
# - Develop a framework !
#
# + [markdown] slideshow={"slide_type": "slide"}
# # DON'T: What teams want to avoid
#
# 1. Avoid sharing too many ideas to the point every team member is building exactly the same solution
| 10,436 |
/Exercise7/exercise7.ipynb | c95567b62bfc8afe4fb63286a78fb6062100e91a | [] | no_license | pintub/ml-coursera-python-assignments | https://github.com/pintub/ml-coursera-python-assignments | 0 | 0 | null | 2020-03-15T17:47:10 | 2020-03-15T08:40:49 | null | Jupyter Notebook | false | false | .py | 1,506,558 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
board = Board()
board.play()
# +
board = Board()
assert not board.is_check('white')
assert not board.is_check('black')
assert not board.is_checkmate('white')
assert not board.is_checkmate('black')
boards = [
Board([
[King('black'), Tower('white')],
[Tower('white'), Queen('white')],
]),
Board([
[King('black'), Horse('white'), Empty()],
[Empty(), Queen('white'), Empty()],
[Empty(), Empty(), Bishop('white')]
]),
Board([
[King('black'), Empty(), Horse('white')],
[Empty(), Empty(), Empty()],
[Horse('white'), Empty(), Bishop('white')]
]),
]
for board in boards:
assert board.is_check('black')
assert board.is_checkmate('black')
boards = [
Board([
[King('black'), Empty(), Horse('white')],
[Empty(), Empty(), Empty()],
[Empty(), Horse('white'), Bishop('white')]
]),
]
for board in boards:
assert not board.is_checkmate('black')
# -
print(board)
# +
import random
random.choice(list(board.moves('white')))
# +
from random import choice
class Opponent:
"""The opponent to play against.
Simulates new boards to find the best new board position.
"""
def __init__(self, board):
self.board = board
self.color = 'black'
self.piece_value = {
'Empty': 0,
'Pawn': 1,
'Horse': 3,
'Bishop': 3,
'Tower': 5,
'Queen': 9,
'King': 1e6,
}
def value(self, board):
total_value = 0
for row in board:
for piece in row:
pv = self.piece_value[type(piece).__name__]
if piece.color == self.color:
total_value += pv
else:
total_value -= pv
return total_value
def simulation(self, board):
board.checkmate = False
try:
while not board.checkmate:
moves = board.moves(board.current_color)
from_ = random.choice([
from_ for from_ in moves if len(moves[from_]) != 0
])
to = random.choice(moves[from_])
board = board.simulate_move(from_, to)
board.update()
except:
print(board)
# -
board = Board()
opponent = Opponent(board)
opponent.simulation(board)
# +
import numpy as np
import copy
from itertools import chain
class Board:
"""The chess board.
The board contains all the pieces and the pieces contains a reference to the board.
"""
def __init__(self, board = None):
if board is None:
self.board = self._create_board()
else:
self.board = board
self.current_color = 'white'
@property
def board(self):
"""Is the board of the correct type etc."""
return self._board
@board.setter
def board(self, value):
for row in value:
for piece in row:
piece.board = self
if isinstance(value, list):
value = np.array(value)
# Run checks
if not isinstance(value, np.ndarray):
raise ValueError('Board should be a numpy array.')
self._board = value
@board.deleter
def board(self):
del self._board
def __getitem__(self, position):
if any(idx < 0 for idx in position):
return None
try:
return self.board[position]
except IndexError:
return None
def __setitem__(self, position, piece):
self.board[position] = piece
def __len__(self):
return 8
def __eq__(self, other):
if isinstance(other, Piece):
return self.board == other
elif isinstance(other, Board):
return bool(self.board == other.board)
return False
def __iter__(self):
return self.board.__iter__()
def __str__(self):
lines = []
for idx, row in enumerate(self):
lines.append(str(8 - idx) + ' ' + ' '.join(map(str, row)))
lines.append(' a b c d e f g h')
return '\n'.join(lines)
def play(self):
self.checkmate = False
while not self.checkmate:
print(self)
print()
str_from = input('Move from: ')
str_to = input('Move to: ')
print()
try:
from_ = self.translate(str_from)
to = self.translate(str_to)
if isinstance(self[from_], Empty):
print("Not a piece...\n")
continue
if self[from_] is None:
print("Not on the board...\n")
continue
if to not in self[from_].moves():
print("Invalid move...\n")
continue
if self[from_].color != self.current_color:
print('Not this players turn...\n')
continue
except:
print("Invalid move...\n")
continue
self.move(from_, to)
def update(self):
self.update_color()
self.update_checkmate()
def translate(self, chess_notation):
letter, number = chess_notation
i = 8 - int(number)
j = dict(zip(
['a', 'b', 'c', 'd', 'e', 'f', 'g', 'h'],
range(len(self))
))[letter]
return i, j
def copy(self):
return copy.deepcopy(self)
def move(self, from_, to):
self[to] = self[from_]
self[from_] = Empty(None, self)
return self
def _create_board(self):
"""Create a chess board."""
return np.array([
self.back_row('white'),
self.pawn_row('white'),
] + [
self.empty_row() for i in range(4)
] + [
self.pawn_row('black'),
self.back_row('black'),
])
def back_row(self, color):
return [
Tower(color, self),
Horse(color, self),
Bishop(color, self),
King(color, self),
Queen(color, self),
Bishop(color, self),
Horse(color, self),
Tower(color, self),
]
def pawn_row(self, color):
return [Pawn(color, self) for j in range(len(self))]
def empty_row(self):
return [Empty(None, self) for j in range(len(self))]
def update_color(self):
self.current_color = self.other_color(self.current_color)
def other_color(self, color):
from_to = {'white': 'black', 'black': 'white'}
return from_to[color]
def moves(self, color):
moves = {}
for row in self.board:
for piece in row:
if piece.color == color:
moves[piece.position] = piece.moves()
return moves
def get_king(self, color):
for row in self.board:
for piece in row:
if (piece.color == color) & isinstance(piece, King):
return piece
raise ValueError(f'Missing king of color {color}')
def king_moves(self, color):
return self.get_king(color).moves()
def update_checkmate(self):
if self.is_checkmate(self.current_color):
self.checkmate = True
print(f"{self.current_color} lost the game...\n")
print(self)
def is_checkmate(self, color):
return (
self.is_check(color)
& self.cant_move_out_of_check(color)
)
def is_check(self, color):
king = self.get_king(color)
enemy_moves = list(chain(
*self.moves(self.other_color(color)).values()
))
return king.position in enemy_moves
def cant_move_out_of_check(self, color):
king = self.get_king(color)
for to in king.moves():
new_board = self.simulate_move(king.position, to)
if not new_board.is_check(color):
return False
return True
def simulate_move(self, from_, to):
new_board = self.copy()
return new_board.move(from_, to)
# -
class Piece:
def __init__(self, color=None, board=None):
self.color = color
self.board = board
@property
def position(self):
i, j = np.where(self.board == self)
return int(i), int(j)
@position.setter
def position(self, value):
self.board.move(
from_=self.position,
to=value
)
def move(self, value):
self.board.move(
from_=self.position,
to=value
)
def moves(self):
"""Should return a list of all moves e.g. [(1,1), (1,2), ..]."""
raise NotImplementedError()
def __str__(self):
"""Return a string representation of the piece."""
if self.color is None:
return ' '
return getattr(self, self.color)
def print_moves(self):
new_board = board.copy()
for move in self.moves():
new_board[move] = '.'
class Empty(Piece):
def moves(self):
return None
class King(Piece):
white = u'♔'
black = u'♚'
def moves(self):
moves = []
i, j = self.position
for di in [-1, 0, 1]:
for dj in [-1, 0, 1]:
new_position = (i + di, j + dj)
if self.board[new_position] is None:
continue
elif self.board[new_position].color == self.color:
continue
moves.append(new_position)
return moves
class Pawn(Piece):
white = u'♙'
black = u'♟'
def moves(self):
i, j = self.position
moves = [(i, j)]
moves.extend(self.forward_move(i, j))
moves.extend(self.attacking_moves(i, j))
return moves
def forward_move(self, i, j):
if self.color == 'white':
moves = [(i + 1, j)]
if i == 1:
moves = [(i + 1, j), (i + 2, j)]
elif self.color == 'black':
moves = [(i - 1, j)]
if i == 6:
moves = [(i - 1, j), (i - 2, j)]
else:
raise ValueError(f"Invalid color {self.color}")
if all(
isinstance(self.board[new_position], Empty)
for new_position in moves
):
return moves
return []
def attacking_moves(self, i, j):
if self.color == 'white':
moves = [(i + 1, j - 1), (i + 1, j + 1)]
moves = filter(
lambda new_position: self.board[new_position] == 'black',
moves
)
elif self.color == 'black':
moves = [(i - 1, j - 1), (i - 1, j + 1)]
moves = filter(
lambda new_position: self.board[new_position] == 'white',
moves
)
else:
raise ValueError(f"Invalid color {self.color}")
return moves
class Tower(Piece):
white = u'♖'
black = u'♜'
def moves(self):
i, j = self.position
moves = [(i, j)]
for di, dj in [(-1, 0), (1, 0), (0, -1), (0, 1)]:
for step in range(1, len(self.board)):
new_position = (i + step * di, j + step * dj)
if self.board[new_position] is None:
break
elif isinstance(self.board[new_position], Empty):
moves.append(new_position)
elif self.board[new_position].color == self.color:
break
elif self.board[new_position].color != self.color:
moves.append(new_position)
break
return moves
class Bishop(Piece):
white = u'♗'
black = u'♝'
def moves(self):
i, j = self.position
moves = [(i, j)]
for di in [-1, 1]:
for dj in [-1, 1]:
for s in range(1, len(self.board)):
new_position = (i + s * di, j + s * dj)
if self.board[new_position] is None:
break
elif isinstance(self.board[new_position], Empty):
moves.append(new_position)
elif self.board[new_position].color == self.color:
break
elif self.board[new_position].color != self.color:
moves.append(new_position)
break
return moves
class Horse(Piece):
white = u'♘'
black = u'♞'
def moves(self):
i, j = self.position
moves = [(i, j)]
for di, dj in [
(-2, -1), (2, 1), (2, -1), (-2, 1),
(-1, -2), (1, 2), (1, -2), (-1, 2),
]:
new_position = (i + di, j + dj)
if self.board[new_position] is None:
continue
elif isinstance(self.board[new_position], Empty):
moves.append(new_position)
elif self.board[new_position].color == self.color:
continue
elif self.board[new_position].color != self.color:
moves.append(new_position)
return moves
class Queen(Piece):
white = u'♕'
black = u'♛'
def moves(self):
i, j = self.position
moves = [(i, j)]
for di in [-1, 0, 1]:
for dj in [-1, 0, 1]:
for s in range(1, len(self.board)):
new_position = (i + s * di, j + s * dj)
if self.board[new_position] is None:
break
elif isinstance(self.board[new_position], Empty):
moves.append(new_position)
elif self.board[new_position].color == self.color:
break
elif self.board[new_position].color != self.color:
moves.append(new_position)
break
return moves
| 14,825 |
/ExposicionSVD.ipynb | fbd52778e2b229fe042a18732e791953112a86ef | [] | no_license | Juanfran23/ExposicionSVD | https://github.com/Juanfran23/ExposicionSVD | 0 | 0 | null | null | null | null | Jupyter Notebook | false | false | .py | 14,198 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Python 3.9.6 64-bit
# name: python3
# ---
# # EX07: Data Wrangling
#
# You will define and use functions that are commonly useful when _wrangling_ data in this exercise. You will frequently need your data to be organized in specific ways in order to perform analysis on it and that organization is rarely exactly the "shape" the data is stored in (such as a CSV table). Data _wrangling_ is the process of loading, converting, and reorganizing data so that you can analyze it.
__author__ = "730395347"
# You will implement the utility functions for this exercise in the `data_utils.py` file found in the `exercises/ex07` directory. As you now know, when you import modules in a running Python program, the module is evaluated only once. Since your Jupyter Notebook _kernel_ is running the entire time you are working on functions in `data_utils.py`, we will use a special extension to automatically reload any changes you make _and save_ in modules you import. The special conventions in the cell below are turning this feature on.
# %reload_ext autoreload
# %autoreload 2
print("Autoreload of imported modules enabled. Be sure to save your work in other modules!")
# Data files will be stored in the `data` directory of the workspace. This Notebook is located in `exercises/ex07` directory. If you think of how to _navigate_ from this directory to the `data` directory, you would need to go "two directories up" and then "into the `data` directory". The constant `DATA_DIRECTORY` defined below uses the convention of two dots to refer to "one directory up", so it is a `str` that references the `data` directory _relative_ to this exercise's directory.
#
# Then, another constant is established referencing the path to the data file you will use to test your functions in this exercise.
DATA_DIRECTORY="../../data"
DATA_FILE_PATH=f"{DATA_DIRECTORY}/nc_durham_2015_march_21_to_26.csv"
# ## Part 0. Reading Data from a Stored CSV File into Memory
#
# In this part of the exercise, you will implement utility functions to read a CSV file from your computer's hard-drive storage into your running program's (Jupyter kernel's) memory. Once in memory, computations over the data set are very fast.
#
# By default, your CSV file is read in row-by-row. Storing these rows as a list of "row" dictionaries is one way of _representing_ tabular data.
#
# ### 0.0) Implement the `read_csv_rows` Function
#
# Complete the implementation of the `read_csv_rows` function in `data_utils.py` and be sure to save your work when making changes in that file _before_ re-evaluating the cell below to test it.
#
# Purpose: Read an entire CSV of data into a `list` of rows, each row represented as `dict[str, str]`.
#
# * Function Name: `read_csv_rows`
# * Parameter:
# 1. `str` path to CSV file
# * Return Type: `list[dict[str, str]]`
#
# Implementation hint: refer back to the code you wrote in lecture on 10/19 for reading a CSV file. We give you the code for this function.
#
# There _should be_ 294 rows and 29 columns read from the `nc_durham_2015_march_21_to_26.csv` stops file. Additionally, the column names should print below those stats.
# +
from data_utils import read_csv_rows
data_rows: list[dict[str, str]] = read_csv_rows(DATA_FILE_PATH)
if len(data_rows) == 0:
print("Go implement read_csv_rows in data_utils.py")
print("Be sure to save your work before re-evaluating this cell!")
else:
print(f"Data File Read: {DATA_FILE_PATH}")
print(f"{len(data_rows)} rows")
print(f"{len(data_rows[0].keys())} columns")
print(f"Columns names: {data_rows[0].keys()}")
# -
# ### 0.1) `column_values` Function
#
# Define and implement this function in `data_utils.py`.
#
# Purpose: Produce a `list[str]` of all values in a single `column` whose name is the second parameter.
#
# * Function Name: `column_values`
# * Parameters:
# 1. `list[dict[str, str]]` - a list of rows representing a _table_
# 2. `str` - the name of the column (key) whose values are being selected
# * Return Type: `list[str]`
#
# Implementation strategy: Establish an empty list to store your column values. Loop through every row in the first parameter. Append the value associated with the key ("column") given as the second parameter to your list of column values. After looping through every row, return the list of column values.
#
# Be sure to save your work before re-evaluating the cell below to test it. With the data loaded in `nc_durham_2015_march_21_to_26.csv`, there should be 294 values in the column. The first five values of the `subject_age` column should be 33, 25, 26, 24, 44.
# +
from data_utils import column_values
subject_age: list[str] = column_values(data_rows, "subject_age")
if len(subject_age) == 0:
print("Complete your implementation of column_values in data_utils.py")
print("Be sure to follow the guidelines above and save your work before re-evaluating!")
else:
print(f"Column 'subject_age' has {len(subject_age)} values.")
print("The first five values are:")
for i in range(5):
print(subject_age[i])
# -
# ### 0.2) `columnar` Function
#
# Define and implement this function in `data_utils.py`.
#
# Purpose: _Transform_ a table represented as a list of rows (e.g. `list[dict[str, str]]`) into one represented as a dictionary of columns (e.g. `dict[str, list[str]]`).
#
# Why is this function useful? Many types of analysis are much easier to perform column-wise.
#
# * Function Name: `columnar`
# * Parameter: `list[dict[str, str]]` - a "table" organized as a list of rows
# * Return Type: `dict[str, list[str]]` - a "table" organized as a dictionary of columns
#
# Implementation strategy: Establish an empty dictionary to the your column-oriented table you are building up to ultimately return. Loop through each of the column names in the first row of the parameter. Get a list of each column's values via your `column_values` function defined previously. Then, associate the column name with the list of its values in the dictionary you established. After looping through every column name, return the dictionary.
# +
from data_utils import columnar
data_cols: dict[str, list[str]] = columnar(data_rows)
if len(data_cols.keys()) == 0:
print("Complete your implementation of columnar in data_utils.py")
print("Be sure to follow the guidelines above and save your work before re-evaluating!")
else:
print(f"{len(data_cols.keys())} columns")
print(f"{len(data_cols['subject_age'])} rows")
print(f"Columns names: {data_cols.keys()}")
# -
# ## Part 1. Selecting ("narrowing down") a Data Table
#
# When working with a data set, it is useful to inspect the contents of the table you are working with in order to both be convinced your analysis is on the correct path and to know what steps to take next with specific column names or values.
#
# In this part of the exercise, you will write some useful utility functions to view the first `N` rows of a column-based table (a function named `head`, referring to the top rows of a table) and another function `select` for producing a simpler data table with only the subset of original columns you care about.
#
# ### Displaying Tabular data with the `tabulate` 3rd Party Library
#
# Reading Python's `str` representations of tabular data, in either representation strategy we used above (list of rows vs. dict of cols), is uncomprehensible for data wrangling. This kind of problem is so common a 3rd party library called `tabulate` is commonly used to produce tables in Jupyter Notebooks. This library was was included in your workspace's `requirements.txt` file at the beginning of the semester, so you should already have it installed!
#
# For a quick demonstration of how the `tabulate` library works, consider this simple demo below. You should be able to evaluate it as is without any further changes and see the tabular representation appear.
# +
from tabulate import tabulate
universities: dict[str, list[str, str]] = {"school": ["UNC", "NCSU", "Duke"], "mascot": ["Rameses", "Wolf", "A Literal Devil"], "founded": ["1789", "1887", "1838"]}
tabulate(universities, universities.keys(), "html")
# -
# ### 1.0) `head` Function
#
# Define and implement this function in `data_utils.py`.
#
# Purpose: Produce a new column-based (e.g. `dict[str, list[str]]`) table with only the first `N` (a parameter) rows of data for each column.
#
# Why: Visualizing a table with hundreds, thousands, or millions of rows in it is overwhelming. You frequently want to just see the first few rows of a table to get a sense you are on the correct path.
#
# * Function name: `head`
# * Parameters:
# 1. `dict[str, list[str]]` - a column-based table of data that _will not be mutated_
# 2. `int` - The number of "rows" to include in the resulting list
# * Return type: `dict[str, list[str]]`
#
# Implementation strategy:
#
# 1. Establish an empty dictionary that will serve as the returned dictionary this function is building up.
# 2. Loop through each of the columns in the first row of the table given as a parameter.
# 1. Inside of the loop, establish an empty list to store each of the first N values in the column.
# 2. Loop through the first N items of the table's column,
# 1. Appending each item to the previously list established in step 2.1.
# 3. Assign the produced list of column values to the dictionary established in step 1.
# 3. Return the dictionary.
#
# Once you have correctly implemented this function and saved your work, you should be able to evaluate the cell below and see the first five rows of the data table presented.
# +
from data_utils import head
data_cols_head: dict[str, list[str]] = head(data_cols, 5)
if len(data_cols_head.keys()) != len(data_cols.keys()) or len(data_cols_head["subject_age"]) != 5:
print("Complete your implementation of columnar in data_utils.py")
print("Be sure to follow the guidelines above and save your work before re-evaluating!")
tabulate(data_cols_head, data_cols_head.keys(), "html")
# -
# ## 1.1) `select` Function
#
# Define and implement this function in `data_utils.py`.
#
# Purpose: Produce a new column-based (e.g. `dict[str, list[str]]`) table with only a specific subset of the original columns.
#
# Why: Many data tables will contain many columns that are not related to the analysis you are trying to perform. _Selecting_ only the columns you care about makes it easier to focus your attention on the problem at hand.
#
# * Function Name: `select`
# * Parameters:
# 1. `dict[str, list[str]]` - a column-based table of data that _will not be mutated_
# 2. `list[str]` - the names of the columns to copy to the new, returned dictionary
# * Return type: `dict[str, list[str]]`
#
# Implementation strategy:
#
# 1. Establish an empty dictionary that will serve as the returned dictionary this function is building up.
# 2. Loop through each of the columns _in the second parameter of the function_
# 1. Assign to the column key of the result dictionary the list of values stored in the input dictionary at the same column
# 3. Return the dictionary produced
#
# Once you have correctly implemented this function, you can run the cell below to visualize the first 10 rows of the table and should only see 2 columns: `subject_race`, `subject_sex`.
# +
from data_utils import select
selected_data: dict[str, list[str]] = select(data_cols, ["subject_race", "subject_sex"])
tabulate(head(selected_data, 9), selected_data.keys(), "html")
# -
# ## 1.2) `concat` Function
#
# Define and implement this function in `data_utils.py`.
#
# Purpose: Produce a new column-based (e.g. `dict[str, list[str]]`) table with two column-based tables combined.
#
# Why: You will often have data from different sources that you may wish to combine in some way to perform an analysis
#
# * Function Name: `concat`
# * Parameters:
# 1. Two `dict[str, list[str]]` - a column-based tables of data that _will not be mutated_
# * Return type: `dict[str, list[str]]`
#
# Implementation strategy:
#
# 1. Establish an empty dictionary that will serve as the returned dictionary this function is building up.
# 2. Loop through each of the columns _in the first parameter of the function_
# 1. Assign to the column key of the result dictionary the list of values stored in the first parameter at the same column
# 3. Loop through each of the columns _in the second parameter of the function_
# 1. If the current column key is already in the result dictionary, add on the list of values stored in the second parameter at the same column
# 2. Otherwise, just assign to the column key of the result dictionary the list of values stored in the second parameter at the same column
# 4. Return the dictionary produced
#
# Once you have correctly implemented this function, you can run the cell below to see 5 stops from March 21st and and 5 stops from March 27th.
# +
from data_utils import concat
additional_table: dict[str, list[str]] = columnar(read_csv_rows(f"{DATA_DIRECTORY}/nc_durham_2015_march_26.csv"))
combined = concat(data_cols_head, additional_table)
tabulate(head(combined, 10), combined.keys(), "html")
# -
# ## Part 2. Simple Analysis
#
# Now that you have some functions to read, transform, and select your data, let's perform some simple analysis! With categorical data, _counting_ the frequency of values is a common first step toward getting an overview of the column.
#
# ### 2.0) `count` Function
#
# Given a `list[str]`, this function will produce a `dict[str, int]` where each key is a unique value in the given list and each value associated is the _count_ of the number of times that value appeared in the input list.
#
# * Function name: `count`
# * Parameter: `list[str]` - list of values to count the frequencies of
# * Return Type: `dict[str, int]` - a dictionary of the counts of each of the items in the input list
#
# Implementation strategy:
#
# 1. Establish an empty dictionary to store your built-up result in
# 2. Loop through each item in the input list
# 1. Check to see if that item has already been established as a key in your dictionary. Try the following boolean conditional: `if <item> in <dict>:` -- replacing `<item>` with the variable name of the current value and `<dict>` with the name of your result dictionary.
# 2. If the item is found in the dict, that means there is already a key/value pair where the item is a key. Increase the value associated with that key by 1 (counting it!)
# 3. If the item is not found in the dict, that means this is the first time you are encountering the value and should assign an initial count of `1` to that key in the result dictionary.
# 3. Return the resulting dictionary.
#
# After you complete this function, you should see the following counts with the included data set when you evaluate the cell below:
#
# ~~~
# race_counts: {'black': 187, 'white': 64, 'hispanic': 38, 'asian/pacific islander': 3, 'other': 1, 'unknown': 1}
# sex_counts: {'female': 111, 'male': 183}
# ~~~
# +
from data_utils import count
race_counts: dict[str, int] = count(selected_data["subject_race"])
print(f"race_counts: {race_counts}")
sex_counts: dict[str, int] = count(selected_data["subject_sex"])
print(f"sex_counts: {sex_counts}")
# -
# ## Bonus Content: Charting with `matplotlib`
#
# Once you have completed the functions above, you have completed the requirements of this exercise. However, you should go ahead and read the code in the cell below and try evaluating it for a fun surprise.
#
# In these cells, we are taking the data from the counts you produced in the cell above and producing bar charts using the popular `matplotlib` 3rd party visualization library. The exact details of what each line does is beyond the scope of your concerns, but I'll bet you can reason through it!
# +
from matplotlib import pyplot as plt
fig, axes = plt.subplots(1, 2, figsize=(12, 5), sharey=True)
fig.suptitle("Traffic Stops in Durham - March 21st through 27th - 2015")
axes[0].set_title("By Race")
axes[0].bar(race_counts.keys(), race_counts.values())
axes[0].tick_params(axis='x', labelrotation = 45)
axes[1].set_title("By Sex")
axes[1].bar(sex_counts.keys(), sex_counts.values())
# -
| 16,425 |
/index.ipynb | 799d7e669a19f50c7f8c0ee7ca411c26d27cb942 | [] | no_license | SydneyHerndon/evaluating-regression-lines-staff | https://github.com/SydneyHerndon/evaluating-regression-lines-staff | 0 | 0 | null | 2019-06-11T20:59:25 | 2018-05-24T16:47:58 | null | Jupyter Notebook | false | false | .py | 21,802 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Evaluating regression lines
# ### Learning Objectives
# * Understand what is meant by the errors of a regression line
# * Understand how to calculate the error at a given point
# * Understand how to calculate RSS and why we use it as a metric to evaluate a regression line
# * Understand the difference between RSS and its variation, the RMSE
# ### Introduction
# So far we have seen how lines and formulas can estimate outputs given an input. We can describe any straight line with two different variables:
#
# * $m$ - the slope of the line, and
# * $b$ - the y-intercept
#
# So far we have been rather fast and loose with choosing a line to estimate our output - we simply drew a line between the first and last points of our data set. Well today, we go further. Here, we take our first step towards **training** our model to match our data.
#
# > The first step in training is to calculate our regression line's **accuracy** -- that is, how well our regression line matches our actual data. Calculating a regression line's accuracy is the topic of this lesson.
#
# In future lessons, we will improve upon our regression line's accuracy, so that it better predicts an output.
# ### Determining Quality
# The first step towards calculating a regression line to predict an output is to calculate how well any regression line matches our data. We need to calculate how accurate our regression line is.
#
# Let's find out what this means. Below we have data that represents the budget and revenue of four shows, with `x` being the budget and `y` being the revenue.
# +
first_show = {'x': 0, 'y': 100}
second_show = {'x': 100, 'y': 150}
third_show = {'x': 200, 'y': 600}
fourth_show = {'x': 400, 'y': 700}
shows = [first_show, second_show, third_show, fourth_show]
shows
# -
# > Run code above with shift + enter
#
# #### An initial regression line
# As we did in the last lab, let's draw a not-so-great regression line simply by drawing a line between our first and last points. We can use our `build_regression_line` function to do so. You can view the code directly [here](https://github.com/learn-co-curriculum/evaluating-regression-lines/blob/master/linear_equations.py).
# > Eventually, we'll improve this regression line. But first we need to see how good or bad a regression line is.
from linear_equations import build_regression_line
x_values = list(map(lambda show: show['x'],shows))
y_values = list(map(lambda show: show['y'],shows))
regression_line = build_regression_line(x_values, y_values)
regression_line
# We can plot our regression line as the following using the [plotting functions](https://github.com/learn-co-curriculum/evaluating-regression-lines/blob/master/graph.py) that we wrote previously:
from graph import m_b_trace, plot, trace_values
from plotly.offline import iplot, init_notebook_mode
init_notebook_mode(connected=True)
data_trace = trace_values(x_values, y_values)
regression_trace = m_b_trace(regression_line['m'], regression_line['b'], x_values)
plot([regression_trace, data_trace])
# So that is what our regression line looks like. And this the line translated into a function.
def sample_regression_formula(x):
return 1.5(x) + 100
# #### Assessing the regression line
#
# Ok, so now that we see what our regression line looks like, let's highlight how well our regression line matches our data.
# 
# > Let's interpret the chart above. That first red line shows that our regression formula does not perfectly predict that first show.
# > * Our actual data -- the first blue dot -- shows that when $x = 100$, $y = 150$.
# > * However, our regression line predicts that at $x = 100$, $y = 250$.
#
# > So **our regression line is off by 100, indicated by the length of the red line.**
# Each point where our regression line's estimated differs from the actual data is called an **error**. And our red lines display the size of this error. The length of the red line equals the size of the error.
# * The **error** equals the difference between the *actual* value and the value *expected* by our model (that is, our regression line).
# * error = actual - expected
#
# Now let's put this formula into practice. The error is the actual value minus the expected value. So at point $x = 100$, the actual $y$ is 150. And at point x = 100, the expected value of $y$ is $250$. So:
# * error = $150 - 250 = -100$.
#
# If we did not have a graph to display this, we could calculate this error by using our formula for the regression line.
#
# * Our regression formula is $y = 1.5x + 100$.
# * Then when $x$ equals 100, the formula predicts $y = 1.5 * 100 + 100 = 250$.
# * And we have the actual data of (100, 150). So
# * `actual` - `expected` $ = 150 -250 = -100$.
# ### Refining our Terms
# Now that we have explained how to calculate an error given a regression line and data, let's learn some mathematical notation that let's us better express these concepts.
#
# * We want to use notation to distinguish between two things: our expected $y$ values and our actual $y$ values.
#
# #### Expected values
#
# So far we have defined our regression function as $y = mx + b$. Where for a given value of $x$, we can calculate the value of $y$. However, this is not totally accurate - as our regression line is not calculating the actual value of $y$ but the *expected* value of $y$. So let's indicate this, by changing our regression line formula to look like the following:
#
# * $\hat{y} = \hat{m}x + \hat{b}$
# Those little dashes over the $y$, $m$ and $b$ are called hats. So our function reads as y-hat equals m-hat multiplied by $x$ plus b-hat. These hats indicate that this formula does not give us the actual value of $y$, but simply our estimated value of $y$. The hats also say that this estimated value of $y$ is based on our estimated values of $m$ and $b$.
# > Note that $x$ is not a predicted value. This is because we are *providing* a value of $x$, not predicting it. For example, we are providing an show's budget as an input, not predicting it. So we are *providing* a value of $x$ and asking it to *predict* a value of $y$.
# #### Actual values
#
# Now remember that we were given some real data as well. This means that we do have actual points for $x$ and $y$, which look like the following.
# +
first_show = {'x': 0, 'y': 100}
second_show = {'x': 100, 'y': 150}
third_show = {'x': 200, 'y': 600}
fourth_show = {'x': 400, 'y': 700}
shows = [first_show, second_show, third_show, fourth_show]
shows
# -
# So how do we represent our actual values of $y$? Here's how: $y$. No extra ink is needed.
#
# Ok, so now we know the following:
# * **$y$**: actual y
# * **$\hat{y}$**: estimated y
#
# Finally, we use the Greek letter $\varepsilon$, epsilon, to indicate error. So we say that
# * $\varepsilon = y - \hat{y}$.
#
# We can be a little more precise by saying we are talking about error at any specific point, where $y$ and $\hat{y}$ are at that $x$ value. This is written as:
#
# $\varepsilon _{i}$ = $y_{i}$ - $\hat{y}_{i}$
# Those little $i$s represent an index value, as in our first, second or third movie. Now, applying this to a specific point of say when $ x = 100 $, we can say:
# * $\varepsilon _{x=100} = y_{x=100}$ - $\hat{y}_{x=100} = 150 - 250 = -100$
# ### Calculating and representing total error
# We now know how to calculate the error at a given value of $x$, $x_i$, by using the formula, $\varepsilon_i$ = $y_i - \hat{y_i}$. Again, this is helpful at describing how well our regression line predicts the value of $y$ at a specific point.
#
# However, we want to see well our regression describes our dataset in general - not just at a single given point. Let's move beyond calculating the error at a given point to describing the total error of the regression line across all of our data.
#
# As an initial approach, we simply calculate the total error by summing the errors, $y - \hat{y}$, for every point in our dataset.
#
# Total Error = $\sum_{i=1}^{n} y_i - \hat{y_i}$
#
# This isn't bad, but we'll need to modify this approach slightly. To understand why, let's take another look at our data.
# 
# The errors at $x = 100$ and $x = 200$ begin to cancel each other out.
#
# * $\varepsilon_{x=100}= 150 - 250 = -100$
# * $\varepsilon_{x=200} = 600 - 400 = 200$
# * $\varepsilon_{x=100} + \varepsilon_{x=200} = -100 + 200 = 100 $
#
# We don't want the errors to cancel each other out! To resolve this issue, we square the errors to ensure that we are always summing positive numbers.
#
# ${\varepsilon_i^2}$ = $({y_i - \hat{y_i}})^2$
#
# So given a list of points with coordinates (x, y), we can calculate the squared error of each of the points, and sum them up. This is called our ** residual sum of squares ** (RSS). Using our sigma notation, our formula RSS looks like:
#
# $ RSS = \sum_{i = 1}^n ({y_i - \hat{y_i}})^2 = \sum_{i = 1}^n \varepsilon_i^2 $
#
# > Residual Sum of Squares is just what it sounds like. A residual is simply the error -- the difference between the actual data and what our model expects. We square each residual and add them together to get RSS.
#
# Let's calculate the RSS for our regression line and associated data. In our example, we have actual $x$ and $y$ values at the following points:
# * $ (0, 100), (100, 150), (200, 600), (400, 700) $.
#
# And we can calculate the values of $\hat{y} $ as $\hat{y} = 1.5 *x + 100 $, for each of those four points. So this gives us:
#
# $RSS = (0 - 0)^2 + (150 - 250)^2 + (600 - 400)^2 + (700 - 700)^2$
#
# which reduces to
#
# $RSS = 0^2 + (-100)^2 + 200^2 + 0^2 = 50,000$
# Now we have one number, the RSS, that represents how well our regression line fits the data. We got there by calculating the errors at each of our provided points, and then squaring the errors so that our errors are always positive.
# ### Root Mean Squared Error
# Root Mean Squared Error, is just a variation on RSS. Essentially, it tries to answer the question of what is the "typical" error of our model versus each data point. To do this, it scales down the size of that large RSS number. So where:
#
# * $ RSS = \sum_{i = 1}^n ({y_i - \hat{y_i}})^2$
#
#
# * $RMSE = \frac{\sqrt{RSS}}{{n}} $
# > Where n equals the number of elements in the data set.
#
# Now let's walk through the reasoning for each step.
#
# #### Taking the square root
# The first thing that makes our RSS large is the fact that we square each error. Remember that we squared each error, because we didn't want positive errors and negative errors to cancel out. Remember, we said that each place where we had a negative error, as in :
# * $actual - expected = -100$,
# * we would square the error, such that $(-100)^2 = 10,000$.
#
# Remember that we square each of our errors, which led to:
#
# * $RSS = 0^2 + (-100)^2 + 200^2 + 0^2 = 50,000$
# With RMSE, after squaring and adding the error we then take the square root of that sum.
# $\sqrt{0^2 + (-100)^2 + 200^2 + 0^2} = \sqrt{50,000} = 223.6$
# #### Taking the Mean
# Now in addition to accounting for the square of each error, RMSE accounts for one other thing as well. Notice that with each additional data point in our data set, our error will tend to increase. So with an increased dataset, RSS will increase. To counteract the effect of RSS increasing with the dataset and not just accuracy, the formula for RMSE divides by the size of the dataset. So continuing along with our above example:
# $ RMSE = \frac{\sqrt{0^2 + (-100)^2 + 200^2 + 0^2}}{4} = \frac{\sqrt{50,000}}{4} = \frac{223.6}{4} = 55.9$
# And generically, we say that:
# $ RMSE = \frac{\sqrt{\sum_{i = 1}^n ({y_i - \hat{y_i}})^2}}{n}$
# So the RMSE gives a typical estimate of how far each measurement is from the expectation. So this is "typical error" as opposed to an overall error.
# ### Summary
# Before this lesson, we simply assumed that our regression line made good predictions of $y$ for given values of $x$. In this lesson, we learned a metric that tells us how well our regression line fits our actual data. To do this, we started looking at the error at a given point, and defined error as the actual value of $y$ minus the expected value of $y$ from our regression line. Then we were able to determine how well our regression line describes the entire dataset by squaring the errors at each point (to eliminate negative errors), and adding these squared errors. This is called the Residual Sum of Squares (RSS). This is our metric for describing how well our regression line fits our data. Lastly, we learned how the RMSE tells us the "typical error" by dividing the square root of the RSS by the number of elements in our dataset.
hat you take the time to learn how to appropriately pre-process tensors for pre-trained models in the [PyTorch documentation](http://pytorch.org/docs/stable/torchvision/models.html).
# **Before writing function**, let us understand how to provide input data to the network, and what kind of output we receive. As usual the documentation is a bit confusing, but on the web there are some resources where they do go around small details for single image processing:
# - We first load the image with Pillow
# - Then we construct the transformation for preprocessing:
# + Resize image
# + Crop the central part
# + Transform it to a tensor
# + Normalize according to documentation
# + Unsquezze the array to copy it to the network
# + Move it to the cuda device
# - We put VGG16 in evaluation mode
# - Then, we evaluate it and take it back to the cpu
# - Finally, we need to detach the tensor to forget about the derivatives and transform it into numpy
# - The largest value is the label
# +
#https://www.learnopencv.com/pytorch-for-beginners-image-classification-using-pre-trained-models/
from PIL import Image
import torchvision.transforms as transforms
img = Image.open(dog_files[0])
normalize = transforms.Normalize(mean = [0.485, 0.456, 0.406],
std = [0.229, 0.224, 0.225])
transform = transforms.Compose([transforms.Resize(256),
transforms.CenterCrop(224),
transforms.ToTensor(),
normalize])
imgtd = transform(img)
batch = torch.unsqueeze(imgtd, 0)
if use_cuda:
batch = batch.cuda()
#torch.utils.data.DataLoader
VGG16.eval()
output = VGG16(batch).cpu()
outputnp = output.detach().numpy()
plt.plot(outputnp.flatten(),'.')
print('Class is:', outputnp.argmax())
# -
# Now, we are ready to write it
# +
from PIL import Image
import torchvision.transforms as transforms
# Set PIL to be tolerant of image files that are truncated.
from PIL import ImageFile
ImageFile.LOAD_TRUNCATED_IMAGES = True
# -
def VGG16_predict(img_path):
'''
Use pre-trained VGG-16 model to obtain index corresponding to
predicted ImageNet class for image at specified path
Args:
img_path: path to an image
Returns:
Index corresponding to VGG-16 model's prediction
'''
## TODO: Complete the function.
## Load and pre-process an image from the given img_path
## Return the *index* of the predicted class for that image
img = Image.open(img_path)
# Preprocessing
# Defining function
normalize = transforms.Normalize(mean = [0.485, 0.456, 0.406],
std = [0.229, 0.224, 0.225])
transform = transforms.Compose([transforms.Resize(256),
transforms.CenterCrop(224),
transforms.ToTensor(),
normalize])
# Transforming
imgtd = transform(img)
batch = torch.unsqueeze(imgtd, 0)
if use_cuda:
batch = batch.cuda()
# Network evaluation
VGG16.eval()
output = VGG16(batch).cpu()
outputnp = output.detach().numpy()
return outputnp.argmax() # predicted class index
# ### (IMPLEMENTATION) Write a Dog Detector
#
# While looking at the [dictionary](https://gist.github.com/yrevar/942d3a0ac09ec9e5eb3a), you will notice that the categories corresponding to dogs appear in an uninterrupted sequence and correspond to dictionary keys 151-268, inclusive, to include all categories from `'Chihuahua'` to `'Mexican hairless'`. Thus, in order to check to see if an image is predicted to contain a dog by the pre-trained VGG-16 model, we need only check if the pre-trained model predicts an index between 151 and 268 (inclusive).
#
# Use these ideas to complete the `dog_detector` function below, which returns `True` if a dog is detected in an image (and `False` if not).
### returns "True" if a dog is detected in the image stored at img_path
def dog_detector(img_path):
## TODO: Complete the function.
imgclass = VGG16_predict(img_path)
isdog = False
if imgclass>150 and imgclass<269:
isdog = True
return isdog # true/false
# ### (IMPLEMENTATION) Assess the Dog Detector
#
# __Question 2:__ Use the code cell below to test the performance of your `dog_detector` function.
# - What percentage of the images in `human_files_short` have a detected dog?
# - What percentage of the images in `dog_files_short` have a detected dog?
# __Answer:__ VGG16 classifies 0% of images of humans as dogs, in this subset, and 99% of images of dogs as dogs. This can be compared to mobilenet v2 (in notebook:data exploration.ipynb), where again 0% of images of humans are classified as dogs and 100% of images of dogs are classified as dogs.
#
# +
# %%time
### TODO: Test the performance of the dog_detector function
### on the images in human_files_short and dog_files_short.
for k, files in enumerate([human_files_short, dog_files_short]):
count = 0
for file in files:
# Is it a dog?
isdog = dog_detector(file)
count+= isdog*1
if k == 0:
print('It classified {}% of human images as dogs'.format(int(count*100/len(human_files_short))))
else:
print('It classified {}% of dogs images as dogs'.format(int(count*100/len(dog_files_short))))
# -
# We suggest VGG-16 as a potential network to detect dog images in your algorithm, but you are free to explore other pre-trained networks (such as [Inception-v3](http://pytorch.org/docs/master/torchvision/models.html#inception-v3), [ResNet-50](http://pytorch.org/docs/master/torchvision/models.html#id3), etc). Please use the code cell below to test other pre-trained PyTorch models. If you decide to pursue this _optional_ task, report performance on `human_files_short` and `dog_files_short`.
# +
### (Optional)
### TODO: Report the performance of another pre-trained network.
### Feel free to use as many code cells as needed
mobilenetv2 = models.mobilenet_v2(pretrained=True)
if use_cuda:
mobilenetv2 = mobilenetv2.cuda()
def mobilenet_predict(img_path):
'''
Use pre-trained mobilenet model to obtain index corresponding to
predicted ImageNet class for image at specified path
Args:
img_path: path to an image
Returns:
Index corresponding to VGG-16 model's prediction
'''
img = Image.open(img_path)
# Preprocessing
# Defining function
normalize = transforms.Normalize(mean = [0.485, 0.456, 0.406],
std = [0.229, 0.224, 0.225])
transform = transforms.Compose([transforms.Resize(256),
transforms.CenterCrop(224),
transforms.ToTensor(),
normalize])
# Transforming
imgtd = transform(img)
batch = torch.unsqueeze(imgtd, 0)
if use_cuda:
batch = batch.cuda()
# Network evaluation
mobilenetv2.eval()
output = mobilenetv2(batch).cpu()
outputnp = output.detach().numpy()
return outputnp.argmax() # predicted class index
#This will override the previous function
def dog_detector(img_path):
imgclass = mobilenet_predict(img_path)
isdog = False
if imgclass>150 and imgclass<269:
isdog = True
return isdog
# -
# **Note**. The reported statistics of mobilenet can be found in data exploration notebook: 0% humans are classified as dogs, and 100% of dogs are classified as dogs. We decided to change to this method since is lighter than VGG, and probably more accurate (although results cannot be distinguished statistically, we would need to test them in a larger dataset, not only 100 samples).
# ---
# <a id='step3'></a>
# ## Step 3: Create a CNN to Classify Dog Breeds (from Scratch)
#
# Now that we have functions for detecting humans and dogs in images, we need a way to predict breed from images. In this step, you will create a CNN that classifies dog breeds. You must create your CNN _from scratch_ (so, you can't use transfer learning _yet_!), and you must attain a test accuracy of at least 10%. In Step 4 of this notebook, you will have the opportunity to use transfer learning to create a CNN that attains greatly improved accuracy.
#
# We mention that the task of assigning breed to dogs from images is considered exceptionally challenging. To see why, consider that *even a human* would have trouble distinguishing between a Brittany and a Welsh Springer Spaniel.
#
# Brittany | Welsh Springer Spaniel
# - | -
# <img src="images/Brittany_02625.jpg" width="100"> | <img src="images/Welsh_springer_spaniel_08203.jpg" width="200">
#
# It is not difficult to find other dog breed pairs with minimal inter-class variation (for instance, Curly-Coated Retrievers and American Water Spaniels).
#
# Curly-Coated Retriever | American Water Spaniel
# - | -
# <img src="images/Curly-coated_retriever_03896.jpg" width="200"> | <img src="images/American_water_spaniel_00648.jpg" width="200">
#
#
# Likewise, recall that labradors come in yellow, chocolate, and black. Your vision-based algorithm will have to conquer this high intra-class variation to determine how to classify all of these different shades as the same breed.
#
# Yellow Labrador | Chocolate Labrador | Black Labrador
# - | -
# <img src="images/Labrador_retriever_06457.jpg" width="150"> | <img src="images/Labrador_retriever_06455.jpg" width="240"> | <img src="images/Labrador_retriever_06449.jpg" width="220">
#
# We also mention that random chance presents an exceptionally low bar: setting aside the fact that the classes are slightly imabalanced, a random guess will provide a correct answer roughly 1 in 133 times, which corresponds to an accuracy of less than 1%.
#
# Remember that the practice is far ahead of the theory in deep learning. Experiment with many different architectures, and trust your intuition. And, of course, have fun!
#
# ### (IMPLEMENTATION) Specify Data Loaders for the Dog Dataset
#
# Use the code cell below to write three separate [data loaders](http://pytorch.org/docs/stable/data.html#torch.utils.data.DataLoader) for the training, validation, and test datasets of dog images (located at `dogImages/train`, `dogImages/valid`, and `dogImages/test`, respectively). You may find [this documentation on custom datasets](http://pytorch.org/docs/stable/torchvision/datasets.html) to be a useful resource. If you are interested in augmenting your training and/or validation data, check out the wide variety of [transforms](http://pytorch.org/docs/stable/torchvision/transforms.html?highlight=transform)!
# +
## Add small gaussian noise to pixel values
class GaussianNoise:
'''This class is another augmentation method that just add
small Gaussian noise to the image. The scale of the noise is
set by the parameter std. Note that if it is additive it should
be behind normalization function, or it can produce negative
pixel values, no clipping for the time being.
'''
def __init__(self, std = 0.02, noisetype = 'additive'):
self.std = std
self.noisetype = noisetype
if self.noisetype == 'additive':
self.transform = lambda x: x + torch.randn(x.size())*std
elif self.noisetype == 'multiplicative':
self.transform = lambda x: x*(1.0+torch.randn(x.size())*std)
else:
print('Noise type not implemented, use either additive or multiplicative. Set to identity')
self.transform = lambda x: x
def __call__(self, x):
return self.transform(x)
# +
import os
from torchvision import datasets
### TODO: Write data loaders for training, validation, and test sets
## Specify appropriate transforms, and batch_sizes
#Hyperparameters
batch_sizes = 8
max_angle = 20
color_r = (0., 0., 0., 0.0) #(0.5, 0.5, 0.001, 0.001)
max_noise = 0.0 #0.01
# Preprocessing
normalize = transforms.Normalize(mean = [0.485, 0.456, 0.406],
std = [0.229, 0.224, 0.225])
data_transform = {'train': transforms.Compose([transforms.Resize(256),
transforms.RandomCrop(224),
transforms.RandomRotation(max_angle),
transforms.RandomVerticalFlip(),
transforms.RandomHorizontalFlip(),
transforms.ColorJitter(brightness=color_r[0],
contrast=color_r[1],
saturation=color_r[2],
hue=color_r[3]),
transforms.RandomPerspective(),
transforms.ToTensor(),
normalize,
GaussianNoise(max_noise, noisetype='additive')
]),
'val': transforms.Compose([transforms.Resize(256),
transforms.CenterCrop(224),
transforms.ToTensor(),
normalize,
]),
'test': transforms.Compose([transforms.Resize(256),
transforms.CenterCrop(224),
transforms.ToTensor(),
normalize,
]),
}
#Datasets
train_data = datasets.ImageFolder('dogImages/train',
transform = data_transform['train'])
valid_data = datasets.ImageFolder('dogImages/valid',
transform = data_transform['val'])
test_data = datasets.ImageFolder('dogImages/test',
transform = data_transform['test'])
data_loaders = {'train': torch.utils.data.DataLoader(train_data,
batch_size = batch_sizes,
shuffle = True,
num_workers = 1
),
'valid':torch.utils.data.DataLoader(valid_data,
batch_size = batch_sizes,
shuffle = True,
num_workers = 1
),
'test':torch.utils.data.DataLoader(test_data,
batch_size = batch_sizes,
shuffle = True,
num_workers = 1
)
}
# -
class_names = train_data.classes
# **Question 3:** Describe your chosen procedure for preprocessing the data.
# - How does your code resize the images (by cropping, stretching, etc)? What size did you pick for the input tensor, and why?
# - Did you decide to augment the dataset? If so, how (through translations, flips, rotations, etc)? If not, why not?
#
# **Answer**:
# This is the chain of transformations:
# - Image is resized to 256x256 pixels to standardize all the iamges
# - A random section of the image of 224x224 pixels is cropped.
# - Applied a rotation with a random angle from $-\theta_\max$ to $\theta_\max$, where $\theta_{\max} = 30$ degrees.
# - Applied randomly a vertical and/or a horizontal flip.
# - Add color jitter in the image, small enough so that it cannot be seen.
# - Normalization of images, using the averages and standard deviations that were used in ImageNet dataset, much larger than our dog dataset.
# - Applied small (additive) Gaussian noise, with standard deviation of 0.05.
#
# We use size 224x224 for the input tensor, so we can use it for mobilenetv2 and other pretrained models. We also decided to augment the dataset as we foresee overfitting issues given the small dataset.
# +
# https://stackoverflow.com/questions/55179282/display-examples-of-augmented-images-in-pytorch
def denormalise(img):
''' It reverses normalization for img, by applying the inverse normalization function.
'''
# transform PIL image to a normal numpy array
img = img.numpy().squeeze().transpose(1, 2, 0)
mean = np.array([0.485, 0.456, 0.406])
std = np.array([0.229, 0.224, 0.225])
img = (img*std + mean).clip(0,1)
return img
sampler = torch.utils.data.DataLoader(train_data,
batch_size = 1,
shuffle = True,
)
fig, axs = plt.subplots(3, 5, figsize = (14,10) )
axs = axs.flatten()
for ax in axs:
img, idx = next(iter(sampler))
ax.imshow(denormalise(img))
ax.set_axis_off()
ax.set_title(class_names[idx], fontsize =9)
fig.subplots_adjust(wspace=0.02, hspace=0)
fig.suptitle('Examples of augmented images')
# -
# ### (IMPLEMENTATION) Model Architecture
#
# Create a CNN to classify dog breed. Use the template in the code cell below.
# +
import torch.nn as nn
import torch.nn.functional as F
# define the CNN architecture
def createblocklist(n0, n1, nf, kernels, nrep = 3, nblocks = 2, pool ='maxpool'):
'''This function return a list of modules to be implemented as a sequential layer block
Input variables:
n0: index in nf array for the layer before this block
n1: index in nf array, for the output filter dimension
nf: array with number of filters (or strides for pooling layers) for different layers of the network
kernels: corresponding arrray of kernels to nf
nrep: number of times layers of convolution+ReLU are repeated in a subblock
nblocks: number of blocks
pool: type of 2d pooling it can be maxpool or anything else that will be considered average pooling.
Output:
list of pytorch layers
'''
# Initial conv+relu that takes previous number of filters to the new one
modlist = [nn.Conv2d(nf[n0],nf[n1]
,kernels[n1], padding = kernels[n1]//2),
nn.ReLU()]
# Chain of nrep conv+relu
for i in range(nrep):
modlist.extend([nn.Conv2d(nf[n1],nf[n1]
,kernels[n1], padding = kernels[n1]//2),
nn.ReLU()]
)
# Only if there are more than one block we do the following block
if nblocks>1:
for j in range(nblocks-1):
# First we compress the number of filters to the initial one
# This might be useful if we to add skip connections later on.
modlist.extend([nn.Conv2d(nf[n1],nf[n0]
,kernels[n1], padding = kernels[n1]//2),
nn.ReLU()]
)
# And decompress it
modlist.extend([nn.Conv2d(nf[n0],nf[n1]
,kernels[n1], padding = kernels[n1]//2),
nn.ReLU()]
)
# Chain of nrep conv+relu
for i in range(nrep):
modlist.extend([nn.Conv2d(nf[n1],nf[n1]
,kernels[n1], padding = kernels[n1]//2),
nn.ReLU()]
)
# Final pooling layer
n1 += 1
if pool =='maxpool':
modlist.append(nn.MaxPool2d(kernels[n1],
stride = nf[n1], padding = kernels[n1]//2)
)
else:
modlist.append(nn.AvgPool2d(kernels[n1],
stride = nf[n1])
)
return modlist
class Net(nn.Module):
''' Add intermediate layers with hyperparameters
as filters and all those things, it will finish with a dense layer
or maybe relu
Input variables:
nf: array with number of filters (or strides for pooling layers) for different layers of the network
kernels: corresponding arrray of kernels to nf
intrablock1repetition : corresponding array of number of times blocks are repeated (unused at the moment)
intrablock2repetition : corresponding array of number of times subblocks are repeated (unused at the moment)
'''
### TODO: choose an architecture, and complete the class
def __init__(self, nf = [32,0,32], kernels = [5,5,5],
intrablock1repetition = [3,3,3],
intrablock2repetition = [3,3,3]):
super(Net, self).__init__()
## Define layers of a CNN
self.layerdict = {'Conv': nn.Conv2d,
'Linear': nn.Linear,
'ReLU' : nn.ReLU,
'maxpool': nn.MaxPool2d,
'avgpool': nn.AvgPool2d
}
self.output_size = len(class_names)
# Arrays of filters (strides for pooling layers) and kernels
self.nf = nf
self.kernels = kernels
self.convlayers = nn.ModuleList([])
n1 = 0
modlist = [nn.Conv2d(3,nf[n1]
,kernels[n1], padding = kernels[n1]//2),
nn.ReLU()]
for i in range(0):
modlist.extend([nn.Conv2d(nf[n1],nf[n1]
,kernels[n1], padding = kernels[n1]//2),
nn.ReLU()]
)
n1 += 1
modlist.append(nn.MaxPool2d(kernels[n1],
stride = nf[n1], padding = kernels[n1]//2)
)
self.convblock0 = nn.Sequential(*modlist)
n0, n1 = 0, 2
modlist = createblocklist(n0, n1, nf, kernels,
nrep = 0, nblocks = 1)
self.convblock1 = nn.Sequential(*modlist)
n0, n1 = 2, 4
modlist = createblocklist(n0, n1, nf, kernels,
nrep = 0, nblocks = 1)
self.convblock2 = nn.Sequential(*modlist)
n0, n1 = 4, 6
modlist = createblocklist(n0, n1, nf, kernels,
nrep = 0, nblocks = 1, pool = 'avgpool')
self.convblock3 = nn.Sequential(*modlist)
# Final two fully-connected layers. Ideally sp_dim should be calculated from previous operations.
sp_dim = 2 # int(224/2**4)
nfF = nf[6]
self.newdim = sp_dim*sp_dim*nfF
self.linear1 = nn.Linear(self.newdim,500)
self.linear2 = nn.Linear(500,self.output_size)
self.dropout = nn.Dropout(0.4)
self.relu = nn.ReLU()
def forward(self, x):
## Define forward behavior
x = self.convblock0(x)
x = self.convblock1(x)
x = self.convblock2(x)
x = self.convblock3(x)
shape = torch.prod(torch.tensor(x.shape[1:])).item()
x = x.view(-1, shape)
x = self.dropout(x)
x = self.linear1(x)
x = self.relu(x)
x = self.linear2(x)
# x = nn.Softmax(dim = 1)(x)
return x
#-#-# You do NOT have to modify the code below this line. #-#-#
# instantiate the CNN
model_scratch = Net(nf = [32,2,64,2,128,2,128,14], kernels = [5,5,5,5,5,5,5,14])
# move tensors to GPU if CUDA is available
if use_cuda:
model_scratch.cuda()
# +
from torchsummary import summary
summary(model_scratch, input_size = (3,224,224))
# -
# __Question 4:__ Outline the steps you took to get to your final CNN architecture and your reasoning at each step.
# __Answer:__ I thought about doing a similar architecture to ResNets, that is why there is a subroutine: createblocklist. Training fairly deep (of around 10 conv+relu layers) I realized very quickly that training this network would be really difficult if we do not include skip connections and batch normalization. The main problem was that the network was not decreasing the training loss. To solve this issue, I decided to return to a very simplified version with only 4 conv+relu with max pooling in between, keeping a small number of parameters, and train it. It happened to start converging after a few attempts with different number of filters.
# ### (IMPLEMENTATION) Specify Loss Function and Optimizer
#
# Use the next code cell to specify a [loss function](http://pytorch.org/docs/stable/nn.html#loss-functions) and [optimizer](http://pytorch.org/docs/stable/optim.html). Save the chosen loss function as `criterion_scratch`, and the optimizer as `optimizer_scratch` below.
# +
import torch.optim as optim
### TODO: select loss function
criterion_scratch = nn.CrossEntropyLoss()
### TODO: select optimizer
optimizer_scratch = optim.SGD(model_scratch.parameters(), lr = .5e-2, momentum = 0.9)
# -
# ### (IMPLEMENTATION) Train and Validate the Model
#
# Train and validate your model in the code cell below. [Save the final model parameters](http://pytorch.org/docs/master/notes/serialization.html) at filepath `'model_scratch.pt'`.
# +
# the following import is required for training to be robust to truncated images
from PIL import ImageFile
ImageFile.LOAD_TRUNCATED_IMAGES = True
loaders_scratch = data_loaders
def train(n_epochs, loaders, model, optimizer, criterion, use_cuda, save_path):
"""returns trained model"""
# initialize tracker for minimum validation loss
valid_loss_min = np.Inf
for epoch in range(1, n_epochs+1):
# initialize variables to monitor training and validation loss
train_loss = 0.0
valid_loss = 0.0
train_acc = 0.0
valid_acc = 0.0
###################
# train the model #
###################
model.train()
for batch_idx, (data, target) in enumerate(loaders['train']):
# move to GPU
if use_cuda:
data, target = data.cuda(), target.cuda()
## find the loss and update the model parameters accordingly
## record the average training loss, using something like
## train_loss = train_loss + ((1 / (batch_idx + 1)) * (loss.data - train_loss))
# Computing the loss
yhat = model(data)
loss = criterion(yhat, target)
# Compute the gradients
loss.backward()
# Update parameters and gradients to zero
optimizer.step()
optimizer.zero_grad()
train_loss +=((1/(batch_idx + 1)) * (loss.data - train_loss))
_, maxidcs = torch.max(yhat,1)
train_acc += (1/(batch_idx + 1))*(-train_acc+
(maxidcs == target).sum().data.cpu().numpy()/maxidcs.size()[0])
del data
del target
######################
# validate the model #
######################
model.eval()
for batch_idx, (data, target) in enumerate(loaders['valid']):
# move to GPU
if use_cuda:
data, target = data.cuda(), target.cuda()
## update the average validation loss
yhat = model(data)
loss = criterion(yhat, target)
valid_loss += ((1/(batch_idx + 1))*(loss.data - valid_loss))
_, maxidcs = torch.max(yhat,1)
valid_acc += (1/(batch_idx + 1))*(-valid_acc+
(maxidcs == target).sum().data.cpu().numpy()/maxidcs.size()[0])
del data
del target
# print training/validation statistics
print('Epoch: {} \tTraining Loss: {:.3f}, acc:{:.3f} \tValidation Loss: {:.6f}, acc:{:.3f}'.format(
epoch,
train_loss,
train_acc,
valid_loss,
valid_acc
))
## TODO: save the model if validation loss has decreased
if valid_loss < valid_loss_min:
torch.save(model.state_dict(),save_path)
# return trained model
return model
# -
model_scratch.load_state_dict(torch.load('model_scratch.pt'))
# +
# train the model
model_scratch = train(10, loaders_scratch, model_scratch, optimizer_scratch,
criterion_scratch, use_cuda, 'model_scratch.pt')
# load the model that got the best validation accuracy
model_scratch.load_state_dict(torch.load('model_scratch.pt'))
# -
optimizer_scratch = optim.SGD(model_scratch.parameters(), lr = .5e-3, momentum = 0.9)
# %%time
model_scratch = train(5, loaders_scratch, model_scratch, optimizer_scratch,
criterion_scratch, use_cuda, 'model_scratch.pt')
# ### (IMPLEMENTATION) Test the Model
#
# Try out your model on the test dataset of dog images. Use the code cell below to calculate and print the test loss and accuracy. Ensure that your test accuracy is greater than 10%.
model_scratch.load_state_dict(torch.load('model_scratch.pt'))
# +
def test(loaders, model, criterion, use_cuda):
# monitor test loss and accuracy
test_loss = 0.
correct = 0.
total = 0.
model.eval()
for batch_idx, (data, target) in enumerate(loaders['test']):
# move to GPU
if use_cuda:
data, target = data.cuda(), target.cuda()
# forward pass: compute predicted outputs by passing inputs to the model
output = model(data)
# calculate the loss
loss = criterion(output, target)
# update average test loss
test_loss = test_loss + ((1 / (batch_idx + 1)) * (loss.data - test_loss))
# convert output probabilities to predicted class
pred = output.data.max(1, keepdim=True)[1]
# compare predictions to true label
correct += np.sum(np.squeeze(pred.eq(target.data.view_as(pred))).cpu().numpy())
total += data.size(0)
print('Test Loss: {:.6f}\n'.format(test_loss))
print('\nTest Accuracy: %2d%% (%2d/%2d)' % (
100. * correct / total, correct, total))
# call test function
test(loaders_scratch, model_scratch, criterion_scratch, use_cuda)
# -
# ---
# <a id='step4'></a>
# ## Step 4: Create a CNN to Classify Dog Breeds (using Transfer Learning)
#
# You will now use transfer learning to create a CNN that can identify dog breed from images. Your CNN must attain at least 60% accuracy on the test set.
#
# ### (IMPLEMENTATION) Specify Data Loaders for the Dog Dataset
#
# Use the code cell below to write three separate [data loaders](http://pytorch.org/docs/master/data.html#torch.utils.data.DataLoader) for the training, validation, and test datasets of dog images (located at `dogImages/train`, `dogImages/valid`, and `dogImages/test`, respectively).
#
# If you like, **you are welcome to use the same data loaders from the previous step**, when you created a CNN from scratch.
# +
## TODO: Specify data loaders
batch_sizes = 8
max_angle = 30
color_r = (0.2, 0.2, 0.001, 0.001)
max_noise = 0.05
path = 'dogImages/'
# Preprocessing
normalize = transforms.Normalize(mean = [0.485, 0.456, 0.406],
std = [0.229, 0.224, 0.225])
data_transform = {'train': transforms.Compose([transforms.Resize(256),
transforms.RandomCrop(224),
transforms.RandomRotation(max_angle),
transforms.RandomVerticalFlip(),
transforms.RandomHorizontalFlip(),
transforms.ColorJitter(brightness=color_r[0],
contrast=color_r[1],
saturation=color_r[2],
hue=color_r[3]),
transforms.ToTensor(),
normalize,
GaussianNoise(max_noise, noisetype='additive')
]),
'val': transforms.Compose([transforms.Resize(256),
transforms.CenterCrop(224),
transforms.ToTensor(),
normalize,
]),
'test': transforms.Compose([transforms.Resize(256),
transforms.CenterCrop(224),
transforms.ToTensor(),
normalize,
]),
}
#Datasets
train_data = datasets.ImageFolder(path+'train',
transform = data_transform['train'])
valid_data = datasets.ImageFolder(path+'valid',
transform = data_transform['val'])
test_data = datasets.ImageFolder(path+'test',
transform = data_transform['test'])
loaders_transfer = {'train': torch.utils.data.DataLoader(train_data,
batch_size = batch_sizes,
shuffle = True,
num_workers = 1
),
'valid':torch.utils.data.DataLoader(valid_data,
batch_size = batch_sizes,
shuffle = True,
num_workers = 1
),
'test':torch.utils.data.DataLoader(test_data,
batch_size = batch_sizes,
shuffle = True,
num_workers = 1
)
}
# -
# ### (IMPLEMENTATION) Model Architecture
#
# Use transfer learning to create a CNN to classify dog breed. Use the code cell below, and save your initialized model as the variable `model_transfer`.
# +
import torchvision.models as models
import torch.nn as nn
## TODO: Specify model architecture
model_transfer = models.mobilenet_v2(pretrained=True)
num_classes = len(class_names)
nft = model_transfer.last_channel
model_transfer.classifier = nn.Sequential(
nn.Dropout(0.2),
nn.Linear(nft, num_classes),
)
if use_cuda:
model_transfer = model_transfer.cuda()
# -
# __Question 5:__ Outline the steps you took to get to your final CNN architecture and your reasoning at each step. Describe why you think the architecture is suitable for the current problem.
# __Answer:__ I tried two different models: Mobilenetv2 and ResNet16. While ResNet16 was training in Udacity’s notebook, Mobilenetv2 was being trained on my laptop. During the training of both algorithms, the training loss of Mobilenetv2 decreased much faster than ResNet16, not only per epoch, but also partially due to the fact it takes less time to train an epoch. Therefore I focused on mobilenet-based architecture
#
# ### (IMPLEMENTATION) Specify Loss Function and Optimizer
#
# Use the next code cell to specify a [loss function](http://pytorch.org/docs/master/nn.html#loss-functions) and [optimizer](http://pytorch.org/docs/master/optim.html). Save the chosen loss function as `criterion_transfer`, and the optimizer as `optimizer_transfer` below.
criterion_transfer = nn.CrossEntropyLoss()
optimizer_transfer = optim.Adam(model_transfer.parameters(), lr = 1e-3)
# ### (IMPLEMENTATION) Train and Validate the Model
#
# Train and validate your model in the code cell below. [Save the final model parameters](http://pytorch.org/docs/master/notes/serialization.html) at filepath `'model_transfer.pt'`.
# train the model
n_epochs = 10
model_transfer = train(n_epochs, loaders_transfer, model_transfer, optimizer_transfer, criterion_transfer, use_cuda, 'model_transfer.pt')
n_epochs = 20
model_transfer = train(n_epochs, loaders_transfer, model_transfer, optimizer_transfer, criterion_transfer, use_cuda, 'model_transfer.pt')
n_epochs = 20
model_transfer = train(n_epochs, loaders_transfer, model_transfer, optimizer_transfer, criterion_transfer, use_cuda, 'model_transfer.pt')
# 36 epochs to reach 0.67 in validation set
n_epochs = 20
model_transfer = train(n_epochs, loaders_transfer, model_transfer, optimizer_transfer, criterion_transfer, use_cuda, 'model_transfer.pt')
# load the model that got the best validation accuracy (uncomment the line below)
model_transfer.load_state_dict(torch.load('model_transfer.pt'))
# ### (IMPLEMENTATION) Test the Model
#
# Try out your model on the test dataset of dog images. Use the code cell below to calculate and print the test loss and accuracy. Ensure that your test accuracy is greater than 60%.
test(loaders_transfer, model_transfer, criterion_transfer, use_cuda)
lossar = np.loadtxt('lossmobilenetv2_f.txt')
fig, ax = plt.subplots(1,2, figsize = (14,5))
ax[0].plot(lossar[:,0],'.-',label='training')
ax[0].plot(lossar[:,2],'.-',label='validation')
ax[0].set_xlabel('epochs', fontsize = 14)
ax[0].set_ylabel('Loss', fontsize = 14)
ax[1].plot(lossar[:,1],'.-',label='training')
ax[1].plot(lossar[:,3],'.-',label='validation')
ax[1].set_xlabel('epochs', fontsize = 14)
ax[1].set_ylabel('Accuracy', fontsize = 14)
ax[0].legend(fontsize = 14)
fig.savefig('loss_mobilenetv2.png', dpi = 200, tight_layout = True)
# ### (IMPLEMENTATION) Predict Dog Breed with the Model
#
# Write a function that takes an image path as input and returns the dog breed (`Affenpinscher`, `Afghan hound`, etc) that is predicted by your model.
# +
### TODO: Write a function that takes a path to an image as input
### and returns the dog breed that is predicted by the model.
# list of class names by index, i.e. a name can be accessed like class_names[0]
#class_names = [item[4:].replace("_", " ") for item in data_transfer['train'].classes]
class_names = [item[4:].replace("_", " ") for item in class_names]
# -
def predict_breed_transfer(img_path):
# load the image and return the predicted breed
model_transfer.eval()
img = Image.open(img_path)
img_transfd = data_transform['test'](img)
batch = torch.unsqueeze(img_transfd, 0)
if use_cuda:
batch = batch.cuda()
output = model_transfer(batch)
outputnp = output.detach().cpu().numpy()
#print(outputnp)
return class_names[outputnp.argmax()]
# %timeit predict_breed_transfer(dog_files_short[50]), dog_files_short[50]
# ---
# <a id='step5'></a>
# ## Step 5: Write your Algorithm
#
# Write an algorithm that accepts a file path to an image and first determines whether the image contains a human, dog, or neither. Then,
# - if a __dog__ is detected in the image, return the predicted breed.
# - if a __human__ is detected in the image, return the resembling dog breed.
# - if __neither__ is detected in the image, provide output that indicates an error.
#
# You are welcome to write your own functions for detecting humans and dogs in images, but feel free to use the `face_detector` and `dog_detector` functions developed above. You are __required__ to use your CNN from Step 4 to predict dog breed.
#
# Some sample output for our algorithm is provided below, but feel free to design your own user experience!
#
# 
#
#
# ### (IMPLEMENTATION) Write your Algorithm
# +
### TODO: Write your algorithm.
### Feel free to use as many code cells as needed.
def run_app(img_path, plot = True):
## handle cases for a human face, dog, and neither
# First we detect what it is:
ishumanface = face_detector(img_path)
isdog = dog_detector(img_path)
if not (ishumanface or isdog):
output = 'Error, no human or dog found in the picture'
else:
if plot:
img = cv2.imread(img_path)
cv_rgb = cv2.cvtColor(img, cv2.COLOR_BGR2RGB)
fig = plt.figure(figsize = (4,4))
ax = fig.add_subplot(111)
ax.imshow(cv_rgb)
ax.set_axis_off()
breed = predict_breed_transfer(img_path)
if not isdog:
#print('Hello you human!')
#print('I would say you look like a {}'.format(breed))
message = 'This human looks like a {}'.format(breed)
output = (0, breed)
else:
#print('This cute dog is a {}'.format(breed))
message = 'This cute dog is a {}'.format(breed)
output = (1, breed)
if plot:
ax.set_title(message)
return output
# -
# ---
# <a id='step6'></a>
# ## Step 6: Test Your Algorithm
#
# In this section, you will take your new algorithm for a spin! What kind of dog does the algorithm think that _you_ look like? If you have a dog, does it predict your dog's breed accurately? If you have a cat, does it mistakenly think that your cat is a dog?
#
# ### (IMPLEMENTATION) Test Your Algorithm on Sample Images!
#
# Test your algorithm at least six images on your computer. Feel free to use any images you like. Use at least two human and two dog images.
#
# __Question 6:__ Is the output better than you expected :) ? Or worse :( ? Provide at least three possible points of improvement for your algorithm.
# __Answer:__
#
# The output is worse. It fails particularly often with Border Collies or Daschunds. In the case of border collies, by inspecting the images one of the reasons is that they only have black and white dogs. With Daschunds it is not clear to me why. One way to improve the algorithm is to provide a more varied dataset, correcting the issues that the previous one has. A second way to improve is to be more aggresive with data augmentation which could extend for a bit longer the period before overfitting. A third way could be using a different approach, by creating a triple GAN, with a generator, a discriminator and a classifier -- this network.
# +
# %%time
## TODO: Execute your algorithm from Step 6 on
## at least 6 images on your computer.
## Feel free to use as many code cells as needed.
new_human_files = ['testimages/fakeGAN.jpg',
'testimages/pablo.jpg',
'testimages/pablo2.jpg']
new_dog_files = ['testimages/002.jpg',
'testimages/001.jpg',
'testimages/003.jpg']
## suggested code, below
for file in np.hstack((new_human_files[:3], new_dog_files[:3])):
run_app(file)
# -
# ### Architecture of Mobilenet v2
summary(model_transfer, input_size = (3,224,224))
| 55,414 |
/dphi_final_bootcamp_car_insurance/notebooks/prepare_data.ipynb | a9b1ab5e1aa7020385a83d433e875a04b22fabc0 | [] | no_license | heavy-data-analysis/data-science-competitions | https://github.com/heavy-data-analysis/data-science-competitions | 0 | 0 | null | null | null | null | Jupyter Notebook | false | false | .py | 508,789 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Python 2
# language: python
# name: python2
# ---
# # Load the modules to used
# +
# %matplotlib inline
print(__doc__)
from time import time
from datetime import datetime
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from sklearn import metrics
from sklearn.cluster import KMeans
from sklearn.datasets import load_digits
from sklearn.decomposition import PCA
from sklearn.preprocessing import scale
path = '/Users/humi/Documents/mobike-intern/Traffic_circles_of_Equal_travel_time/DATA/'
all_order = pd.DataFrame()
# -
# # Data cleaning
# ## a)CBD
# +
f = open(path + "dengshiquan/CBD","r")
lines = f.readlines()#读取全部内容
lines_drop_track = []
for line in lines:
lines_drop_track.append(line.split("===")[0])
starttime = [line.split(",")[1] for line in lines_drop_track][1:]
startpositionx = [line.split(",")[2] for line in lines_drop_track][1:]
startpositiony = [line.split(",")[3] for line in lines_drop_track][1:]
endtime = [line.split(",")[4] for line in lines_drop_track][1:]
endpositionx = [line.split(",")[5] for line in lines_drop_track][1:]
endpositiony = [line.split(",")[6] for line in lines_drop_track][1:]
df1 = pd.DataFrame()
df1['startpositionx'] = startpositionx
df1['startpositiony'] = startpositiony
df1['endpositionx'] = endpositionx
df1['endpositiony'] = endpositiony
difftime = []
for i in range(df1.shape[0]):
st = datetime.strptime(str(starttime[i]),"%Y-%m-%d %H:%M:%S.%f")
ed = datetime.strptime(str(endtime[i]),"%Y-%m-%d %H:%M:%S.%f")
difftime.append((ed-st).seconds/60)
df1['time'] = difftime
#df1.to_csv(path + 'DATA_CLEANED/CBD.csv',encoding='UTF-8')
# -
data = df1[['startpositionx','startpositiony']]
kmeans = KMeans(init='k-means++', n_clusters=5, n_init=10)
kmeans.fit(data)
Z = kmeans.predict(data)
centroids = kmeans.cluster_centers_
centroids
# ## b)Jinrongjie
# +
f = open(path + "dengshiquan/jingrongjie","r")
lines = f.readlines()#读取全部内容
lines_drop_track = []
for line in lines:
lines_drop_track.append(line.split("===")[0])
starttime = [line.split(",")[1] for line in lines_drop_track][1:]
startpositionx = [line.split(",")[2] for line in lines_drop_track][1:]
startpositiony = [line.split(",")[3] for line in lines_drop_track][1:]
endtime = [line.split(",")[4] for line in lines_drop_track][1:]
endpositionx = [line.split(",")[5] for line in lines_drop_track][1:]
endpositiony = [line.split(",")[6] for line in lines_drop_track][1:]
df2 = pd.DataFrame()
df2['startpositionx'] = startpositionx
df2['startpositiony'] = startpositiony
df2['endpositionx'] = endpositionx
df2['endpositiony'] = endpositiony
difftime = []
for i in range(df2.shape[0]):
st = datetime.strptime(str(starttime[i]),"%Y-%m-%d %H:%M:%S.%f")
ed = datetime.strptime(str(endtime[i]),"%Y-%m-%d %H:%M:%S.%f")
difftime.append((ed-st).seconds/60)
df2['time'] = difftime
#df2.to_csv(path + 'DATA_CLEANED/jinrongjie.csv',encoding='UTF-8')
# -
df2['time'].values.min()
# ## c)Wangjing
# +
f = open(path + "dengshiquan/wangjing","r")
lines = f.readlines()#读取全部内容
lines_drop_track = []
for line in lines:
lines_drop_track.append(line.split("===")[0])
starttime = [line.split(",")[1] for line in lines_drop_track][1:]
startpositionx = [line.split(",")[2] for line in lines_drop_track][1:]
startpositiony = [line.split(",")[3] for line in lines_drop_track][1:]
endtime = [line.split(",")[4] for line in lines_drop_track][1:]
endpositionx = [line.split(",")[5] for line in lines_drop_track][1:]
endpositiony = [line.split(",")[6] for line in lines_drop_track][1:]
df3 = pd.DataFrame()
df3['startpositionx'] = startpositionx
df3['startpositiony'] = startpositiony
df3['endpositionx'] = endpositionx
df3['endpositiony'] = endpositiony
difftime = []
for i in range(df3.shape[0]):
st = datetime.strptime(str(starttime[i]),"%Y-%m-%d %H:%M:%S.%f")
ed = datetime.strptime(str(endtime[i]),"%Y-%m-%d %H:%M:%S.%f")
difftime.append((ed-st).seconds/60)
df3['time'] = difftime
# df3.to_csv(path + 'DATA_CLEANED/wangjing.csv',encoding='UTF-8')
# -
# ## d)Zhongguancun
# +
f = open(path + "dengshiquan/zhongguancun","r")
lines = f.readlines()#读取全部内容
lines_drop_track = []
for line in lines:
lines_drop_track.append(line.split("===")[0])
starttime = [line.split(",")[1] for line in lines_drop_track][1:]
startpositionx = [line.split(",")[2] for line in lines_drop_track][1:]
startpositiony = [line.split(",")[3] for line in lines_drop_track][1:]
endtime = [line.split(",")[4] for line in lines_drop_track][1:]
endpositionx = [line.split(",")[5] for line in lines_drop_track][1:]
endpositiony = [line.split(",")[6] for line in lines_drop_track][1:]
df4 = pd.DataFrame()
df4['startpositionx'] = startpositionx
df4['startpositiony'] = startpositiony
df4['endpositionx'] = endpositionx
df4['endpositiony'] = endpositiony
difftime = []
for i in range(df4.shape[0]):
st = datetime.strptime(str(starttime[i]),"%Y-%m-%d %H:%M:%S.%f")
ed = datetime.strptime(str(endtime[i]),"%Y-%m-%d %H:%M:%S.%f")
difftime.append((ed-st).seconds/60)
df4['time'] = difftime
#df4.to_csv(path + 'DATA_CLEANED/zhongguancun.csv',encoding='UTF-8')
# -
# # e)Xierqi
# +
f = open(path + "dengshiquan/xierqi","r")
lines = f.readlines()#读取全部内容
lines_drop_track = []
for line in lines:
lines_drop_track.append(line.split("===")[0])
starttime = [line.split(",")[1] for line in lines_drop_track][1:]
startpositionx = [line.split(",")[2] for line in lines_drop_track][1:]
startpositiony = [line.split(",")[3] for line in lines_drop_track][1:]
endtime = [line.split(",")[4] for line in lines_drop_track][1:]
endpositionx = [line.split(",")[5] for line in lines_drop_track][1:]
endpositiony = [line.split(",")[6] for line in lines_drop_track][1:]
df5 = pd.DataFrame()
df5['startpositionx'] = startpositionx
df5['startpositiony'] = startpositiony
df5['endpositionx'] = endpositionx
df5['endpositiony'] = endpositiony
difftime = []
for i in range(df5.shape[0]):
st = datetime.strptime(str(starttime[i]),"%Y-%m-%d %H:%M:%S.%f")
ed = datetime.strptime(str(endtime[i]),"%Y-%m-%d %H:%M:%S.%f")
difftime.append((ed-st).seconds/60)
df5['time'] = difftime
# df5.to_csv(path + 'DATA_CLEANED/xierqi.csv',encoding='UTF-8')
# -
# ## f)Merge data
frames = [df1,df2,df3,df4,df5]
result = pd.concat(frames)
result = result[result['endpositionx'] != 'null']
result = result[result['endpositiony'] != 'null']
# +
distance = []
for i in range(result.shape[0]):
distance.append(haversine(float(result.startpositionx.values[i]),
float(result.startpositiony.values[i]),
float(result.endpositionx.values[i]),
float(result.endpositiony.values[i])))
result['distance'] = distance
# -
speed = result['distance']/result['time']
ulimit = np.percentile(speed.values, 98)
result = result[speed<=ulimit]
result.head()
# # Cluster center
f = open(path + "cluster_center","r")
lines = f.readlines()#读取全部内容
cluster_center = lines[0].split(";")
# # Coordinate of circles of Equal travel time
# +
from math import radians, cos, sin, asin, sqrt
def haversine(lon1, lat1, lon2, lat2): # 经度1,纬度1,经度2,纬度2 (十进制度数)
"""
Calculate the great circle distance between two points
on the earth (specified in decimal degrees)
"""
# 将十进制度数转化为弧度
lon1, lat1, lon2, lat2 = map(radians, [lon1, lat1, lon2, lat2])
# haversine公式
dlon = lon2 - lon1
dlat = lat2 - lat1
a = sin(dlat/2)**2 + cos(lat1) * cos(lat2) * sin(dlon/2)**2
c = 2 * asin(sqrt(a))
r = 6371 # 地球平均半径,单位为公里
return c * r * 1000
# -
for j in range(28):
status = []
for i in range(result.shape[0]):
status.append(haversine(float(result.startpositionx.values[i]),
float(result.startpositiony.values[i]),
float(cluster_center[j].split(",")[1]),
float(cluster_center[j].split(",")[0]))<300)
sub_result = result[status]
sub_result = sub_result.reset_index()[['endpositiony','endpositionx','time']]
sub_result.columns = ['lat','lon','time']
sub_result = sub_result[sub_result.time<200]
sub_result = sub_result.sort_values(by='time')
sub_result.to_csv(path + "RESULT/sub_result_" + str(j) + ".csv",index=False)
result
path + "RESULT/sub_result_" + str(1) + ".csv"
result
len(cluster_center)
| 8,668 |
/MLTIPS/07_handle_unknown_categories.ipynb | 5212003ed5cbe40974885bf96cb351c704bcb8fe | [] | no_license | shivams289/Kaggle-Projects | https://github.com/shivams289/Kaggle-Projects | 0 | 0 | null | null | null | null | Jupyter Notebook | false | false | .py | 6,321 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# [](https://mybinder.org/v2/gh/justmarkham/scikit-learn-tips/master?filepath=notebooks%2F07_handle_unknown_categories.ipynb)
#
# [](https://colab.research.google.com/github/justmarkham/scikit-learn-tips/blob/master/notebooks/07_handle_unknown_categories.ipynb)
#
# # 🤖⚡ scikit-learn tip #7 ([video](https://www.youtube.com/watch?v=bA6mYC1a_Eg&list=PL5-da3qGB5ID7YYAqireYEew2mWVvgmj6&index=7))
#
# Q: For a one-hot encoded feature, what can you do if new data contains categories that weren't seen during training?
#
# A: Set handle_unknown='ignore' to encode new categories as all zeros.
#
# See example 👇
#
# P.S. If you know all possible categories that might ever appear, you can instead specify the categories manually. handle_unknown='ignore' is useful specifically when you don't know all possible categories.
import pandas as pd
X = pd.DataFrame({'col':['A', 'B', 'C', 'B']})
X_new = pd.DataFrame({'col':['A', 'C', 'D']})
from sklearn.preprocessing import OneHotEncoder
ohe = OneHotEncoder(sparse=False, handle_unknown='ignore')
X
# three columns represent categories A, B, and C
ohe.fit_transform(X[['col']])
# category D was not learned by OneHotEncoder during the "fit" step
X_new
# category D is encoded as all zeros
ohe.transform(X_new[['col']])
# ### Want more tips? [View all tips on GitHub](https://github.com/justmarkham/scikit-learn-tips) or [Sign up to receive 2 tips by email every week](https://scikit-learn.tips) 💌
#
# © 2020 [Data School](https://www.dataschool.io). All rights reserved.
| 1,912 |
/08-lists.ipynb | d0c0a5c7f4312ac2d62d6fb7988a537883aacffe | [
"BSD-3-Clause"
] | permissive | trailmarkerlib/exploreDataPython | https://github.com/trailmarkerlib/exploreDataPython | 0 | 0 | null | null | null | null | Jupyter Notebook | false | false | .py | 19,884 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Chapter 8 Lists
# ### 8.1 A list is a sequence
# +
# list of integers
[10, 20, 30, 40]
# list of strings
['frog','toad','salamander','newt']
# mixed list
[10,'twenty',30.0,[40, 45]]
# -
cheeses = ['Cheddar','Mozzarella','Gouda','Swiss']
numbers = [27, 42]
empty = []
print(cheeses, numbers, empty)
# ### 8.2 Lists are mutable
numbers = [27, 62]
numbers[1] = 42
print(numbers)
cheeses = ['Cheddar','Mozzarella','Gouda','Swiss']
'Swiss' in cheeses
'Brie' in cheeses
# ### 8.3 Traversing a list
cheeses = ['Cheddar','Mozzarella','Gouda','Swiss']
for cheese in cheeses:
print(cheese)
numbers = [10, 20, 30, 40]
for i in range(len(numbers)):
numbers[i] = numbers[i] + 2
print(numbers)
empty = []
for x in empty:
print('This never happens.')
len(['spam', 1, ['sun','moon','stars'], [1,2,3]])
# ### 8.4 List operations
a = [1,2,3]
b = [4,5,6]
c = a + b
print(c)
[0]*4
[1,2,3]*3
# ### 8.5 List slices
t = ['a','b','c','d','e','f']
t[1:3]
t[:4]
t[3:]
t[:]
t[1:3] = ['x','y']
print(t)
# ### 8.6 List methods
t = ['a','b','c']
t.append('d')
print(t)
t1 = ['a','b','c']
t2 = ['d','e']
t1.extend(t2)
print(t1)
t = ['d','c','e','b','a']
t.sort()
print(t)
# ### 8.7 Deleting elements
t = ['a','b','c']
x = t.pop(1)
print(t)
print(x)
t = ['a','b','c']
del t[1]
print(t)
t = ['a','b','c']
x = t.remove('b')
print(t)
t = ['a','b','c','b']
x = t.remove('b') #removes first instance
print(t)
t = ['a', 'b', 'c', 'd', 'e','f']
del t[1:5]
print(t)
# ### 8.8 Lists and functions
nums = [3, 41, 12, 9, 74, 15]
print(len(nums))
print(max(nums))
print(min(nums))
print(sum(nums))
print(sum(nums)/len(nums))
numlist = list()
while(True):
inp = input('Enter a number: ')
if inp == 'done': break
try:
value = float(inp)
numlist.append(value)
except:
print('That was not a number. Continue...')
continue
average = sum(numlist)/len(numlist)
print('Average:', average)
# ### 8.9 Lists and strings
s = 'spam'
t = list(s)
print(t)
s = 'That there’s some good in this world, Mr. Frodo… and it’s worth fighting for.'
t = s.split()
print(t)
print(t[3])
s = 'spam-spam-spam'
delimiter = '-'
s.split(delimiter)
t = ['That', 'there’s', 'some', 'good', 'in', 'this', 'world,', 'Mr.', 'Frodo…', 'and', 'it’s', 'worth', 'fighting', 'for.']
delimiter = ' '
delimiter.join(t)
# ### 8.10 Parsing lines
fhand = open('mbox-short.txt')
for line in fhand:
line = line.rstrip()
if not line.startswith('From '): continue
words = line.split()
print(words[2])
# ### 8.11 Objects and values
# same object, two variables pointing to the same object
a = 'banana'
b = 'banana'
a is b
# same value because a and b point to the same object
a == b
# two separate objects
a = [1,2,3]
b = [1,2,3]
a is b
# same value
a == b
# ### 8.12 Aliasing
# b points to the same object as a
a = [1,2,3]
b = a
b is a
b[0] = 17
print(a)
# ### 8.13 List arguments
def delete_head(t):
del t[0]
letters = ['a','b','c']
delete_head(letters)
print(letters)
t1 = [1,2]
t2 = t1.append(3)
print(t1)
print(t2)
t1 = [1,2]
t3 = t1 + [3]
print(t3)
# changes t within the scope of the function but not
# the value of the variable passed to it
def bad_delete_head(t):
t = t[1:]
def tail(t):
return t[1:]
letters = ['a','b','c']
rest = tail(letters)
print(rest)
| 3,607 |
/02-Data_Analysis/2.1 - Pandas Deepdive.ipynb | 9e4756ebb4eefb88789ac331ea1e2e2f4f9851f7 | [] | no_license | carolinaruizlopez/ds_tb_part_21_09 | https://github.com/carolinaruizlopez/ds_tb_part_21_09 | 0 | 0 | null | null | null | null | Jupyter Notebook | false | false | .py | 420,568 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: clase
# language: python
# name: clase
# ---
# # 2.1 - Pandas Deepdive
#
# 
# ### Importar un archivo
import numpy as np # importar librerias
import pandas as pd
pd.read_csv('https://raw.githubusercontent.com/justmarkham/DAT8/master/data/drinks.csv').head()
# +
df=pd.read_excel('../data/Online Retail.xlsx') # cargar archivo
df.head()
# +
df['NullID']=[i if i%2 else None for i in range(len(df))] # columna de nulos
df.head()
# +
# renombrar columnas
df.columns
# +
n_columnas=[c.lower() for c in df.columns]
n_columnas
# +
df.columns=n_columnas
df.head()
# +
df=df.rename(columns={'invoiceno': 'no'})
df.head()
# -
list(zip(df.columns, n_columnas))
# +
df=df.rename(columns={k:v for k,v in zip(df.columns, n_columnas)}) # k=vieja columna, v=nueva columna
df.head()
# -
# ### Descripción de los datos
df.shape
df.info()
df.info(memory_usage='deep')
df.count() # valores no nulos
df['country'].value_counts() # cuenta de valores unicos en la columna
df['country'].unique() # valores unicos en la columna
df['country'].nunique()
len(df['country'].unique())
# +
# descripcion estadistica
df.describe()
# -
df.describe().T
df.describe().T['50%'] # es la mediana
df.median()
# ### Selección dentro del dataframe
df[['quantity', 'unitprice']].head() # seleccion por nombre de columna
df[df.quantity==6].head() # seleccion segun condicion
df[~(df.quantity==6)].head() # ~ es el NO logico, lo que no es 6
df[(df.quantity==6) & (df.unitprice==2.55)].head() # & es el y logico
df[(df.quantity==6) | (df.unitprice==2.55)].head() # | es el o logico
df[~(df.quantity==6) & (df.unitprice==2.55)].head()
df[(df.quantity!=6) & (df.unitprice==2.55)].head()
# +
# seleccion por indice
df.iloc[3]
# -
df.iloc[:2]
df.iloc[:2, 1:3]
df.loc[:2]
df.loc[:2, 'description']
df.loc[:2, ['description', 'country', 'nullid']]
df.sample(4)
df.sample(4, random_state=42) # random state es la semilla de los nos aleatorios
df.sample(frac=0.1).head()
df._get_numeric_data().head()
df.select_dtypes(include=['float64', 'object']).head()
# ### Cambio del tipo de datos
df.quantity.astype(dtype='float32')
# +
df.quantity=df.quantity.astype(dtype='float32')
df.head()
# -
# ### Manejo de valores nulos
df.info()
df.isna().head()
df.notna().head()
# +
nan_cols=df.isna().sum()
nan_cols[nan_cols>0]
# -
# **¿Qué hacemos?**
# +
# rellenar con 0
df.fillna(0).head()
# +
# rellenar con media
df.fillna(df.nullid.mean()).head()
# -
# ### https://www.geeksforgeeks.org/python-pandas-dataframe-ffill/
#
# ### https://pandas.pydata.org/docs/reference/api/pandas.DataFrame.interpolate.html
# +
# rellenar con total
df.fillna(df.nullid.sum()).head()
# +
# rellenar con minimmo
df.fillna(df.nullid.min()).head()
# +
# rellenar con mediana
df.fillna(df.nullid.median()).head()
# +
# borrar columna
df.drop('nullid', axis=1).head()
# -
df.drop(columns=['revenue', 'customerid', 'country']).head()
df[df['nullid'].notna()]
df.dropna().head()
df.drop_duplicates().head()
df.drop_duplicates(subset=['country']).head()
# ### Agrupación
df.groupby('country').agg('count').head() # el groupby necesita siempre siempre una funcion de agragacion
df.groupby('country').agg('sum').head()
df.groupby('country').sum().head()
df.groupby('country').agg({'quantity': 'sum', 'unitprice':'mean'}).head()
df.groupby(['country', 'unitprice']).sum().head()
df.groupby(['country', 'unitprice']).sum().index[:5]
def suma(x):
return sum(x)
df.groupby('country').agg({'quantity': suma}).head()
df.groupby('country').agg({'quantity': 'sum'}).head()
df.groupby('country').agg({'unitprice': ['mean', 'std', 'min', 'max', 'median']}).head()
df.groupby('country').agg(['mean', 'std', 'min', 'max', 'median']).head()
tmp_df=df.groupby('country').agg(['mean', 'std', 'min', 'max', 'median'])
tmp_df['invoiceno']['mean'][:4]
# ### Pivot table
pd.set_option('display.max_columns', None) # para que printee todas las columnas
# +
student=pd.read_csv('../data/student-por.csv', sep=';')
student.head()
# -
student.pivot_table(index=['school'], columns=['sex'])
# +
# %%time
student.pivot_table(index=['school'], columns=['sex'], values=['G3'])
# -
student.pivot_table(index=['school'], columns=['sex'], values=['G3'], aggfunc='mean')
student.pivot_table(index=['school'], columns=['sex'], values=['G3'], aggfunc='count')
student.pivot_table(index=['school'], columns=['sex', 'studytime'], values=['G3'], aggfunc='count')
help(student.pivot_table)
# ### Cross table
pd.crosstab(index=student.sex, columns='count')
# +
# %%time
pd.crosstab(index=student.sex, columns=student.activities)
# -
pd.crosstab(index=student.sex, columns=student.activities, values=student.absences, aggfunc='count')
pd.crosstab(index=student.sex, columns=student.activities, values=student.absences, aggfunc='sum')
help(pd.crosstab)
# ### Aplicación de funciones
# +
# map=mathematical application ; map(funcion, iterable)
# -
def suma_2(x):
return x+2
df.quantity[:5]
list(map(suma_2, df.quantity))[:5]
list(map(suma_2, [1,2,3,4,5]))
# +
# %%time
res=[]
for e in df.quantity:
suma=suma_2(e)
res.append(suma)
df.quantity=res
# +
# %%time
df.quantity=[suma_2(e) for e in df.quantity]
# +
# %%time
df.quantity=list(map(suma_2, df.quantity))
# -
df.head()
df.quantity.apply(suma_2)[:5] # apply es el map de pandas
df.quantity.apply(lambda x: x+2)
df._get_numeric_data().dropna().apply(lambda x: x+2).head()
df['description'].apply(lambda x: x.replace('HEART T-LIGHT HOLDER', 'hola que tal hola que pasa que tal'))[:5]
# ### Json Normalize, Explode
# +
df_json=pd.read_json('../data/companies.json', lines=True)
df_json.head()
# -
df_json.shape
pd.json_normalize(df_json._id)[:5]
pd.json_normalize(df_json.image.dropna()).available_sizes[0]
# ### https://pandas.pydata.org/pandas-docs/version/1.2.0/reference/api/pandas.json_normalize.html
df_json.shape
exp_df=df_json.explode('products')
exp_df.head()
exp_df.shape
# ### Unión de dataframes
df.shape
# +
# concatenación
pd.concat([df, df]).head()
# -
len(pd.concat([df, df]))
pd.concat([df, df], axis=0).head() # condicion necesaria, mismo numero de columnas y nombre
pd.concat([df, df], axis=1).head() # mismo numero de filas
pd.concat([df, df], axis=1).shape
# +
df2=df.copy()
df2.index=[i for i in range(0, 2*len(df), 2)]
df2.head()
# -
pd.concat([df, df2], axis=1).head()
pd.concat([df, df2], axis=1).shape
# +
# join
uno=pd.DataFrame({'key': ['K0', 'K1', 'K2', 'K3', 'K4', 'K5'],
'A': ['A0', 'A1', 'A2', 'A3', 'A4', 'A5']})
otro=pd.DataFrame({'key': ['K0', 'K1', 'K2'],
'B': ['B0', 'B1', 'B2']})
# -
uno.head()
otro.head()
uno.join(otro, lsuffix='_uno', rsuffix='_otro')
uno.set_index('key').join(otro.set_index('key'))
uno.join(otro.set_index('key'), on='key') # left join
uno.join(otro.set_index('key'), on='key', how='right') # right join
# +
# merge
df1=pd.DataFrame({'lkey': ['foo', 'bar', 'baz', 'foo'],
'value': [1, 2, 3, 5]})
df2=pd.DataFrame({'rkey': ['foo', 'bar', 'baz', 'foo'],
'value': [5, 6, 7, 8]})
# -
df1.head()
df2.head()
df1.merge(df2)
df1.merge(df2, left_on='lkey', right_on='rkey', suffixes=('_left', '_right'))
# ### Eval y query
df1=pd.DataFrame(np.random.random((10, 3)), columns=['a', 'b', 'c'])
df2=pd.DataFrame(np.random.random((10, 3)), columns=['a', 'b', 'c'])
df1.head()
df2.head()
pd.eval('df1 > df2').head()
df1.eval('b>0')
df1.query('b > 0.5')
df1.query('b > 0.5 & a > 0.3')
df1.query('b > 0.5 | a > 0.3')
df1.query('b > 0.5 | a > 0.3')['a'].apply(lambda x: x+9)
# ### lookup
seasons=pd.DataFrame(np.random.random((6, 4)),
columns=['winter', 'spring', 'summer', 'autumn'])
seasons
look = ['summer', 'winter', 'summer', 'spring', 'autumn', 'spring']
seasons['look'] = seasons.lookup(seasons.index, look)
seasons.head()
# ### get
seasons.get(seasons.winter>0.5) # devuelveme todo el df donde winter>0.5
# ### index y reindex
seasons.index
seasons.index=pd.RangeIndex(0, 6*2, 2)
seasons.index
seasons.index=range(6)
seasons.index
# +
seasons.index=seasons.look
seasons.head()
# +
seasons.set_index('winter', inplace=True)
seasons.head()
# -
# ### Multi index
# +
seasons.set_index(['look', 'summer'], inplace=True)
seasons.head()
# -
seasons.index
# ### Exportar datos
df.to_csv('../data/retail.csv', index=False)
df.to_json('../data/retail.json', orient='records')
df.to_excel('../data/retail.xlsx', index=False)
df.to_excel('../data/retail.xlsx', 'Sheet2', index=False)
# ### Métodos habituales de pandas
# ```python
# df.head() # printea la cabeza, por defecto 5 filas
# df.tail() # pritea la cola, por defecto 5 filas
# df.describe() # descripcion estadistica
# df.info() # informacion del df
# df.info(memory_usage='deep')
# df.columns # muestra columna
# df.index # muestra indice
# df.dtypes # muestra tipos de datos de las columnas
# df.plot() # hace un grafico
# df.hist() # hace un histograma
# df.col.value_counts() # cuenta los valores unicos de una columna
# df.col.unique() # muestra valores unicos de una columna
# df.copy() # copia el df
# df.drop() # elimina columnas o filas (axis=0,1)
# df.dropna() # elimina nulos
# df.fillna() # rellena nulos
# df.shape # dimensiones del df
# df._get_numeric_data() # selecciona columnas numericas
# df.rename() # renombre columnas
# df.str.replace() # reemplaza columnas de strings
# df.astype(dtype='float32') # cambia el tipo de dato
# df.iloc[] # localiza por indice
# df.loc[] # localiza por elemento
# df.transpose() # transpone el df
# df.T
# df.sample(n, frac) # muestra del df
# df.col.sum() # suma de una columna
# df.col.max() # maximo de una columna
# df.col.min() # minimo de una columna
# df[col] # selecciona columna
# df.col
# df.isnull() # valores nulos
# df.isna()
# df.notna() # valores no nulos
# df.drop_duplicates() # eliminar duplicados
# df.reset_index(inplace=True) # resetea el indice y sobreescribe
# ```
lc0Pbed" outputId="7471cf6a-2ae6-4ec3-ed6b-27f312cc7aca"
# subset_emoji_only_df_extract_emoji = subset_emoji_only_df.withColumn('extracted_emoji', extract_emoji_udf(col('description')))
# tkn_extracted_emoji = RegexTokenizer().setInputCol('extracted_emoji').setOutputCol('emoji_list').setPattern(' ')
# subset_emoji_only_df_extract_emoji = tkn_extracted_emoji.transform(subset_emoji_only_df_extract_emoji)
counts = clean_category_df.select(F.explode('emoji_list').alias('col')).groupBy('col').count().collect()
clean_list = [(row['col'], row['count'] )for row in counts]
emoji_frequency_df = spark.createDataFrame(clean_list , ['emoji','count'])
emoji_frequency_df.orderBy(col('count').desc()).show(5)
# + [markdown] colab_type="text" id="IFbY401gQhYM"
# ## 2.3 Which are the top three most popular emoji categories
# + colab={} colab_type="code" id="E7nHIxwRQl9D"
def assign_category(emo):
if emo in tranportation_emoji:
return 'transportation'
elif emo in people_emoji:
return 'people'
elif emo in food_emoji:
return 'food'
elif emo in event_emoji:
return 'event'
elif emo in activity_emoji:
return 'activity'
elif emo in travel_emoji:
return 'travel'
elif emo in utility_emoji:
return 'utility'
else:
return 'others'
# + colab={"base_uri": "https://localhost:8080/", "height": 187} colab_type="code" id="D4r0lBJfQpJT" outputId="beb41aa9-85fd-415b-c7b6-3640b6a03d4b"
assign_category_udf = F.udf(lambda row:assign_category(row))
emoji_frequency_df_with_category = emoji_frequency_df.withColumn('category', assign_category_udf(col('emoji')))
# Because there are some emojis not in the emoji dictionary, we show 4 most popular categories including 'other' category
emoji_frequency_df_with_category.groupBy('category').sum('count').orderBy(col('sum(count)').desc()).show(4)
# + [markdown] colab_type="text" id="dK6TsfVHOu1F"
# # Question 3: User Spending Profile
# + colab={} colab_type="code" id="6YRf75NIhv7Q"
clean_category_df.show(5)
# + colab={} colab_type="code" id="Y5KY-Lp6he7Y"
# We assume one transaction can possibly belong to two categories, so we extract them out if there's any.
transaction_df = clean_category_df.select(col("user1").alias('User'),'datetime','story_id',
F.explode("clean_category").alias("clean_category"))
# + colab={} colab_type="code" id="dE4F4qjshfC_"
transaction_df.createOrReplaceTempView("transaction_cate")
# + colab={} colab_type="code" id="3kJGcMEkhfGS"
trans_profile = spark.sql("""SELECT User,
SUM(CASE WHEN T.clean_category = 'activity' THEN 1 ELSE 0 END)/count('story_id') AS activity,
SUM(CASE WHEN T.clean_category = 'cash' THEN 1 ELSE 0 END)/count('story_id') AS cash,
SUM(CASE WHEN T.clean_category = 'event' THEN 1 ELSE 0 END)/count('story_id') AS event,
SUM(CASE WHEN T.clean_category = 'food' THEN 1 ELSE 0 END)/count('story_id') AS food,
SUM(CASE WHEN T.clean_category = 'illegal' THEN 1 ELSE 0 END)/count('story_id') AS illegal,
SUM(CASE WHEN T.clean_category = 'people' THEN 1 ELSE 0 END)/count('story_id') AS people,
SUM(CASE WHEN T.clean_category = 'transportation' THEN 1 ELSE 0 END)/count('story_id') AS transportation,
SUM(CASE WHEN T.clean_category = 'travel' THEN 1 ELSE 0 END)/count('story_id') AS travel,
SUM(CASE WHEN T.clean_category = 'utility' THEN 1 ELSE 0 END)/count('story_id') AS utility
from transaction_cate T
group by User""")
# + colab={"base_uri": "https://localhost:8080/", "height": 204} colab_type="code" id="SpCTa3shhfJD" outputId="e60ec510-ec6c-436e-b841-4755b22a5d74"
trans_profile.show(5)
# + [markdown] colab_type="text" id="tMvRhk2AjOxt"
# # Q4: User’s dynamic spending profile
# + colab={} colab_type="code" id="r5DgHOEFh6_6"
transaction_df.show(5)
# + colab={} colab_type="code" id="SIL_EX6Rh7C1"
# Calculate the number of days passed wince the user's first transaction
window = Window.partitionBy('User')
trans_with_daysPassed = transaction_df.withColumn("daysPassed", F.datediff("dateTime", F.min("dateTime").over(window)))
# + colab={} colab_type="code" id="-UXdCC8th7Hr"
# Transfer daysPassed to number of month the user is currently in
assign_month = F.udf(lambda x: 0 if (x == 0) else math.ceil(x/30))
category_with_month = trans_with_daysPassed.withColumn('month', assign_month(F.col('daysPassed')))
category_with_month = category_with_month.withColumn("month", category_with_month["month"].cast(IntegerType()))
# + colab={} colab_type="code" id="904CEM5Qh7Me"
category_with_month.show(5)
# + colab={} colab_type="code" id="2sv3w3Uhh7Pg"
# Since we only consider 0-12 months. We filter the other data to increase the computing efficiency
category_with_month.createOrReplaceTempView("category_with_month")
cate_month_12 = spark.sql("""SELECT * FROM category_with_month WHERE month <= 12""")
# + colab={} colab_type="code" id="yQYAyIOxh7KW"
# Calculate the number of transactions for each user and each month in each category
cate_month_12.createOrReplaceTempView("cate_month_12")
# + colab={} colab_type="code" id="dJFzY6bgh7F1"
profile_month = spark.sql("""SELECT User, month,
count(story_id) AS total_trans,
SUM(CASE WHEN T.clean_category = 'activity' THEN 1 ELSE 0 END) AS activity_num,
SUM(CASE WHEN T.clean_category = 'cash' THEN 1 ELSE 0 END) AS cash_num,
SUM(CASE WHEN T.clean_category = 'event' THEN 1 ELSE 0 END) AS event_num,
SUM(CASE WHEN T.clean_category = 'food' THEN 1 ELSE 0 END) AS food_num,
SUM(CASE WHEN T.clean_category = 'illegal' THEN 1 ELSE 0 END) AS illegal_num,
SUM(CASE WHEN T.clean_category = 'people' THEN 1 ELSE 0 END) AS people_num,
SUM(CASE WHEN T.clean_category = 'transportation' THEN 1 ELSE 0 END) AS transportation_num,
SUM(CASE WHEN T.clean_category = 'travel' THEN 1 ELSE 0 END) AS travel_num,
SUM(CASE WHEN T.clean_category = 'utility' THEN 1 ELSE 0 END) AS utility_num
from cate_month_12 T
group by User, month""")
# + colab={} colab_type="code" id="Ljq42oUXhfQs"
profile_month = profile_month.orderBy('User','month',ascending=False)
# + colab={} colab_type="code" id="JJOYGpSkiWSE"
# Calculate the cumulative number of transactions for each user, month and category
profile_month.createOrReplaceTempView("profile_month")
window = Window.partitionBy('User').orderBy(profile_month["month"].asc()).rowsBetween(Window.unboundedPreceding, Window.currentRow)
# + colab={} colab_type="code" id="oFZlLNrniWVP"
dynamic_profile = profile_month.withColumn("total_sum", sum("total_trans").over(window))\
.withColumn("activity", sum("activity_num").over(window))\
.withColumn("cash", sum("cash_num").over(window))\
.withColumn("event", sum("event_num").over(window))\
.withColumn("food", sum("food_num").over(window))\
.withColumn("illegal", sum("illegal_num").over(window))\
.withColumn("people", sum("people_num").over(window))\
.withColumn("transportation", sum("transportation_num").over(window))\
.withColumn("travel", sum("travel_num").over(window))\
.withColumn("utility", sum("utility_num").over(window))\
# + colab={} colab_type="code" id="H15D_icIiWYf"
# Calculate the percentage
dynamic_profile = dynamic_profile.withColumn("activity", F.col("activity")/F.col('total_sum'))\
.withColumn("cash", F.col("cash")/F.col('total_sum'))\
.withColumn("event", F.col("event")/F.col('total_sum'))\
.withColumn("food", F.col("food")/F.col('total_sum'))\
.withColumn("illegal", F.col("illegal")/F.col('total_sum'))\
.withColumn("people", F.col("people")/F.col('total_sum'))\
.withColumn("transportation", F.col("transportation")/F.col('total_sum'))\
.withColumn("travel", F.col("travel")/F.col('total_sum'))\
.withColumn("utility", F.col("utility")/F.col('total_sum'))
# + colab={} colab_type="code" id="xdWr1cL8iWPz"
dynamic_profile.show()
# + colab={} colab_type="code" id="n12cYH5UijqW"
dynamic_profile_clean = dynamic_profile.drop(*dynamic_profile.columns[3:12])
# + colab={} colab_type="code" id="9KA1BWy2ijx4"
dynamic_profile_clean.show(5)
# + colab={} colab_type="code" id="D8cxkd27ldQh"
dynamic_profile_clean.createOrReplaceTempView("df4")
# + colab={} colab_type="code" id="7sSZ0ago-wUD"
dynamic_profile_clean.createOrReplaceTempView("dynamic_profile_clean")
# + [markdown] colab_type="text" id="_87QRxtulWk7"
# ## Plotting spending profile of the average user
# + colab={} colab_type="code" id="OaLfHBjBij51"
# Save and read the csv to save time
# Don't run it if you have the csv
dynamic_profile_clean.toPandas().to_csv('dynamic_profile_clean.csv')
# + colab={} colab_type="code" id="4vkS4pH0ij8f"
dynamic_profile_clean = spark\
.read\
.option('header','True')\
.option('inferSchema','true')\
.csv(r'dynamic_profile_clean.csv')
# + colab={} colab_type="code" id="KKywZyRJij3a"
dynamic_profile_clean.createOrReplaceTempView("dynamic_profile_clean")
# + colab={} colab_type="code" id="B1HzDYIZi531"
def get_mean_std(cate):
dynamic_profile_gt0 = dynamic_profile_clean.where(cate + ' > 0.0')
dynamic_profile_mean_std = dynamic_profile_gt0.groupBy('month').agg(F.avg(col(cate)).alias('avg'),\
F.stddev(col(cate)).alias('std')).sort(col("month").asc())
return dynamic_profile_mean_std
# + colab={} colab_type="code" id="upHozOAsi57v"
cate_list = dynamic_profile_clean.columns[6:15]
# + colab={} colab_type="code" id="BV-TzsWGi5_B"
def plot_mean_std(df,cate):
plt.plot(df["month"], df["avg"])
plt.fill_between(df["month"], (df["avg"]-2*df["std"]), (df["avg"]+2*df["std"]), color='b', alpha=.1)
plt.title('Average spending profile for ' + cate)
plt.xlabel("month")
plt.ylabel("Average Percentage")
plt.xticks(np.arange(0, 13, step = 1))
# + colab={"base_uri": "https://localhost:8080/", "height": 1000} colab_type="code" id="dFrLRQpPjDKC" outputId="a76d9e28-0267-40c8-889b-c53c09f4ae53"
fig, ax = plt.subplots(figsize = (15,30))
for (i, cate) in zip(range(len(cate_list)), cate_list):
profile_mean_std = get_mean_std(cate)
df = profile_mean_std.toPandas()
plt.subplot(5,2,i+1)
plot_mean_std(df, cate)
# + [markdown] colab_type="text" id="ywyYEtvVLhtR"
# # Social Network Analytics
# + [markdown] colab_type="text" id="ty7R2CZvLhzK"
# ## [5 pts]: Write a script to find a user’s friends and friends of friends (Friend definition:
#
# A user’s friend is someone who has transacted with the user, either sending money to the user or
# receiving money from the user). __Describe your algorithm and calculate its computational
# complexity. Can you do it better?__
# + colab={} colab_type="code" id="if7Zpd2NL8H9"
friend_to = transaction.orderBy('user1', 'user2')
# + colab={} colab_type="code" id="xReAVFzPMA7H"
def rename_columns(df, columns):
'''
This function is used to rename multiple columns in pyspark columns
ARG:
- df: a pysaprk dataframe
- columns: a dict that stores {'replace_column': new_name}
'''
if isinstance(columns, dict):
for old_name, new_name in columns.items():
df = df.withColumnRenamed(old_name, new_name)
return df
else:
raise ValueError("'columns' should be a dict, like {'old_name_1':'new_name_1', 'old_name_2':'new_name_2'}")
# + colab={} colab_type="code" id="H2G4BrbYMBkc"
# friend_from
friend_from = transaction.select('user2', 'user1', 'transaction_type', 'datetime', 'description', 'is_business', 'story_id')\
.orderBy('user2', 'user1')
friend_from1 = rename_columns(friend_from, {'user1': 'user3', 'user2': 'user1', 'user3': 'user2'})
friend_from2 = friend_from1.withColumn('transaction_type',
F.when(friend_from1['transaction_type'] == 'charge', 'payment').
otherwise('charge'))
# + colab={} colab_type="code" id="vNDJDcePMJXM"
# friend_all unions friend_to and friend_from, which allows every transaction to appear twice in the dataframe
# With both users appear at 'user1' column
friend_all = friend_to.union(friend_from2)
# + [markdown] colab_type="text" id="GlK76CNNL481"
# Keep only user_id in the friend_user dataframes for computation convenience
# + colab={} colab_type="code" id="BLq7-Aa8MY5M"
friend_user = friend_all.select('user1', 'user2').orderBy('user1', 'user2')
friend_user1 = friend_user.alias('friend_user1')
friend_user2 = friend_user.alias('friend_user2')
friend_user2 = rename_columns(friend_user2, {'user2': 'friend_of_user2'})
# + [markdown] colab_type="text" id="QfWuUEO5Lhw9"
# We considered using _cond = [f.col("friend_user1.user2") == f.col("friend_user2.user1"), f.col("friend_user1.user1") != f.col("friend_of_user2")]_ to specify that the friend's friends do not contain the user him/herself. However, excluding the user in the friend's friends list won't bring huge effects to the result, and would complicate the algorithm much more, and the effect. So we dicided to keep the original user.
# + colab={} colab_type="code" id="cx4ef-N5L4LS"
# add constraints to join the friends' friends to the table containing user id and his/her friends
cond = [F.col("friend_user1.user2") == F.col("friend_user2.user1")]
friend_friend = friend_user1.join(friend_user2, cond, "inner")\
.drop(F.col("friend_user2.user1"))
friend_friend = friend_friend.orderBy('friend_user1.user1', 'user2', 'friend_of_user2')
# + colab={} colab_type="code" id="WTxJezaHMn8e"
friend_friend_clean = friend_friend.dropDuplicates()
# + colab={} colab_type="code" id="l1W_BzDRMq8b"
friend_friend_clean.show(10)
# + [markdown] colab_type="text" id="_N8mPOMJMv9R"
# 
# + colab={} colab_type="code" id="Y2XZax5XMrX0"
def find_user_friend(user_id):
return friend_all.filter(friend_friend.user1 == user_id).dropDuplicates().select('user1', 'user2')
# + colab={} colab_type="code" id="ooc7jGmsMzXr"
find_user_friend(2).show()
# + [markdown] colab_type="text" id="OY0_XJhLM00N"
# 
# + colab={} colab_type="code" id="IRZvNR08M3wU"
def find_user_friend_and_fof(user_id):
return friend_friend_clean.filter(friend_friend.user1 == user_id).select('user1', 'user2', 'friend_of_user2')
# + colab={} colab_type="code" id="nlGSRaxMM34j"
find_user_friend_and_fof(2).show()
# + [markdown] colab_type="text" id="ILaLRxJ3M0q0"
# 
# + [markdown] colab_type="text" id="5aQV1KTMLhrG"
# ### Number of friends and number of friends of friends
# + colab={} colab_type="code" id="dfYdqDscND-X"
# Calculate the number of days passed wince the user's first transaction
assign_month = F.udf(lambda x: 0 if (x == 0) else math.ceil(x/30))
# + colab={} colab_type="code" id="dQCo_68GNEJM"
# assign the 'month' to user1 and user2
window_user1 = Window.partitionBy('user1')
friend_all_daysPassed = friend_all.withColumn("daysPassed", F.datediff("datetime", F.min("datetime").over(window_user1)))
category_with_user1month = friend_all_daysPassed.withColumn('month', assign_month(F.col('daysPassed')))
category_with_user1month = category_with_user1month.withColumn("month", category_with_user1month["month"].cast(IntegerType()))
window_user2 = Window.partitionBy('user2')
category_with_user2month = category_with_user1month.withColumn("daysPassed2", F.datediff("datetime", F.min("datetime").over(window_user2)))
category_with_month = category_with_user2month.withColumn('month2', assign_month(F.col('daysPassed2')))
category_with_month = category_with_month.withColumn("month2", category_with_month["month2"].cast(IntegerType()))
category_with_month.createOrReplaceTempView("category_with_month")
# + [markdown] colab_type="text" id="np7mW5BpNS3i"
# Calculate the number of friends
# + colab={} colab_type="code" id="SqEjsjOtNESp"
friend_num = spark.sql("""SELECT user1, month, COUNT(DISTINCT user2) AS FRIEND_NUM
FROM category_with_month t1
WHERE month <= 12
GROUP BY user1, month
ORDER BY user1, month""")
friend_num.createOrReplaceTempView("friend_num")
# + [markdown] colab_type="text" id="MZorg-NpNadK"
# Calculate the number of friends' friends
# + colab={} colab_type="code" id="K0NwtXPfNXbM"
# attach the number of friends' of user2 to the huge transaction table
category_with_month_wfof = spark.sql("""SELECT t1.*, t2.friend_num
FROM category_with_month t1
INNER JOIN friend_num t2
ON t1.user2 = t2.user1
AND t1.month2 = t2.month
ORDER BY user1, user2""")
category_with_month_wfof.createOrReplaceTempView("category_with_month_wfof")
# + colab={} colab_type="code" id="jjMugTJZNrSa"
# With user1 and month1 specified, Select only distinct user2 (user1's friends)
fof_num_tmp = spark.sql("""SELECT DISTINCT user1, month, user2, friend_num
FROM category_with_month_wfof
WHERE month <= 12
ORDER BY user1, month""")
fof_num_tmp.createOrReplaceTempView("fof_num_tmp")
# + colab={} colab_type="code" id="PlYO_VqpNrxD"
# join the result
fof_num = spark.sql("""SELECT t1.user1, t1.month, IFNULL(SUM(t2.friend_num), 0) AS fof_num
FROM friend_num t1
LEFT JOIN fof_num_tmp t2
ON t1.user1 = t2.user1
AND t1.month = t2.month
WHERE t1.month <= 12
GROUP BY t1.user1, t1.month
ORDER BY t1.user1, t1.month""")
fof_num.createOrReplaceTempView("fof_num")
# + [markdown] colab_type="text" id="9Ok2dj9hNwP9"
# Combine the number of friends and friends' friends
# + colab={} colab_type="code" id="zj5LsDdXNvSm"
user_f_fof_num = spark.sql("""SELECT t1.user1, t1.month, t1.friend_num, t2.fof_num
FROM friend_num t1
LEFT JOIN fof_num t2
ON t1.user1 = t2.user1
AND t1.month = t2.month
WHERE t1.month <= 12
ORDER BY t1.user1, t1.month""")
user_f_fof_num.createOrReplaceTempView("user_f_fof_num")
# + colab={} colab_type="code" id="qxz8DQ7KN1y7"
# store the results to parquet form
user_f_fof_num.write.parquet('user_f_fof_num.parquet')
# + [markdown] colab_type="text" id="XkK59_rrOl6Q"
# ### Clustering coefficient of a user's network.
# + colab={} colab_type="code" id="PKecOodXOpz7"
# Calculate possible triplets
possible_tri = spark.sql("""
SELECT t1.user1, t1.month,
CASE
WHEN t1.friend_num <= 1 THEN 0
ELSE INT(t1.friend_num * (t1.friend_num -1)/2)
END AS possible_tri
FROM friend_num t1
WHERE t1.month <= 12
ORDER BY t1.user1, t1.month""")
possible_tri.createOrReplaceTempView("possible_tri")
# + colab={} colab_type="code" id="Mj7o1Qk-Pg9D"
# Calculate existed triplets
existed_tri = spark.sql("""
SELECT t1.user1, t1.month, IFNULL(COUNT(DISTINCT *), 0) AS existed_tri
FROM category_with_month t1
INNER JOIN category_with_month t2
ON t1.month <= 12
AND t1.user2 = t2.user1
LEFT JOIN (SELECT DISTINCT t02.user1, t02.month, t02.user2
FROM category_with_month t02
WHERE t02.month <= 12
) t02
ON t02.user2 = t2.user2
AND t02.user1 = t1.user1
AND t02.month = t1.month
GROUP BY t1.user1, t1.month
ORDER BY t1.user1, t1.month""")
existed_tri.createOrReplaceTempView("existed_tri")
# + colab={} colab_type="code" id="2AIqTNTBPmQH"
# Calculate clustering coefficients
cluster_coef = spark.sql("""
SELECT t1.user1, t1.month, IFNULL(existed_tri/possible_tri, 0) AS exicluster_coef
FROM possible_tri t2
LEFT JOIN existed_tri t1
ON t1.user1 = t2.user1
AND t1.month = t2.month
ORDER BY t1.user1, t1.month""")
cluster_coef.createOrReplaceTempView("cluster_coef")
# + colab={} colab_type="code" id="A-dio1LcPrMW"
cluster_coef.show(5)
# + [markdown] colab_type="text" id="DL3FU2SOOmAz"
# 
# + [markdown] colab_type="text" id="j6rocF2JPyKt"
# Save the results from network analytics
# + colab={} colab_type="code" id="l02Dwn96PxGu"
network_analytic = spark.sql("""
SELECT t1.*, IFNULL(t2.exicluster_coef, 0)AS cluster_coef
FROM user_f_fof_num t1
LEFT JOIN cluster_coef t2
ON t1.user1 = t2.user1
AND t1.month = t2.month
ORDER BY t1.user1, t1.month""")
network_analytic.createOrReplaceTempView("network_analytic")
# + colab={} colab_type="code" id="X72rUXPCQlFa"
network_analytic.write.parquet('network_analytic.parquet')
# + [markdown] colab_type="text" id="t0RsIxGaQoos"
# ### Calculate the page rank of each user
#
# _It's a dumb way to calculate the page rank, and probably would take forever to generate the results. But before I think of something smart, I'll leave them here._
# + colab={} colab_type="code" id="0Dd3NOQrQ6oM"
category_with_month_lt12_payment = category_with_month.filter(category_with_month.month <= 12).select('user1', 'month', 'user2', 'month2', 'transaction_type').orderBy('user1', 'month')
category_with_month_lt12_payment = category_with_month_lt12_payment.filter(category_with_month_lt12_payment.transaction_type == 'payment')
# + colab={} colab_type="code" id="W8qSOBseREPW"
user_month = category_with_month_lt12_payment.select\
(F.concat(F.col("user1"), F.lit("_"), F.col("month")),\
F.concat(F.col("user2"), F.lit("_"), F.col("month2")),\
category_with_month_lt12_payment.transaction_type)
user_month = user_month.withColumnRenamed('concat(user1, _, month)', 'user1_month')
user_month = user_month.withColumnRenamed('concat(user2, _, month2)', 'user2_month')
# + colab={} colab_type="code" id="uhQ_8xxpRG22"
user_month_edge = user_month.withColumnRenamed('user1_month', 'src')
user_month_edge = user_month_edge.withColumnRenamed('user2_month', 'dst')
user_month_edge = user_month_edge.withColumnRenamed('transaction_type', 'relationship')
# + colab={} colab_type="code" id="sdJkcBTARIFO"
verticesDf = user_month.select('user1_month').union(user_month.select('user2_month'))
verticesDf = verticesDf.withColumnRenamed('user1_month', 'id')
# + colab={} colab_type="code" id="7oRA55NrRKD2"
g = GraphFrame(verticesDf, user_month_edge)
pr = g.pageRank(resetProbability=0.15, tol=0.01)
# + [markdown] colab_type="text" id="EPwBESADkhzA"
# ## Predictive Analysis with MLlib
# + [markdown] colab_type="text" id="hqtbhqWz6D9u"
# ### Q7. Compute total number of transactions within 12 months for each user
# + colab={} colab_type="code" id="6wn95HwVmV0Z"
from pyspark.sql.functions import *
from pyspark.sql.types import *
# + colab={} colab_type="code" id="2BRUOsNtpWLp"
# create table containing 'user' and the number of transactions within 12 months
transaction.createOrReplaceTempView("transaction2")
df1 = spark.sql("select user1 as user, datetime from transaction2 union all select user2 as user, datetime from transaction2")
df1.createOrReplaceTempView("df1")
df2 = spark.sql("select user, datetime, min(datetime) over (partition by user order by user, datetime) as first_time_transaction from df1")
df2.createOrReplaceTempView("df2")
df3 = spark.sql("select user, date_format(first_time_transaction,'Y/M/d') as first_time_transaction,date_format(datetime,'Y/M/d') as datetime from df2")
df3.createOrReplaceTempView("df3")
df4 = spark.sql("select * from (select user, months_between(datetime, first_time_transaction) as months_after_first_transaction from df2) t where t.months_after_first_transaction < 12")
df4.createOrReplaceTempView("df4")
df5 = spark.sql("select user, count(*) as number_of_transactions_within_12_months from df4 group by user")
df5.createOrReplaceTempView("df5")
# -
df5.show(5)
# + [markdown] colab_type="text" id="465PXHRS6tC-"
# ### Q8. Create the Recency and Frequency variables
# +
from pyspark.sql.window import Window
import pyspark.sql.functions as F
import math
window = Window.partitionBy('User')
trans_with_daysPassed = df1.withColumn("daysPassed", F.datediff("dateTime", F.min("dateTime").over(window)))
assign_month = F.udf(lambda x: 0 if (x == 0) else math.ceil(x/30))
category_with_month = trans_with_daysPassed.withColumn('month', assign_month(F.col('daysPassed')))
category_with_month = category_with_month.withColumn("month", category_with_month["month"].cast(IntegerType()))
# -
category_with_month.createOrReplaceTempView("category_with_month")
cate_month_12 = spark.sql("""SELECT * FROM category_with_month WHERE month <= 12""")
cate_month_12.createOrReplaceTempView("cate_month_12")
cate_month_12.show(10)
# create lifetime table for all users containing t0 - t11
lst = [0,1,2,3,4,5,6,7,8,9,10,11,12]
df_time = df5.withColumn("time_lst", F.array([F.lit(x) for x in lst]))
df_time = df_time.withColumn("time_lst", F.explode(col("time_lst")))
# +
df_sum = spark.sql("select user, month as lifetime_indicator, count(*) as freq_times, \
max(daysPassed) as max_days \
from cate_month_12 \
group by user, month")
df_time.createOrReplaceTempView("df_time")
df_sum.createOrReplaceTempView("df_sum")
# -
df_sum.show()
df_time.show()
# +
df_sum2 = spark.sql("select user, lifetime_indicator, max(max_days) as max_days, \
max(freq_times) as freq_times \
from \
(select user, lifetime_indicator, max_days, freq_times from df_sum \
union \
select user, time_lst, null as max_days, null as freq_times \
from df_time) T \
group by user, lifetime_indicator \
order by user, lifetime_indicator ")
df_sum2.createOrReplaceTempView("df_sum2")
df_sum2.show()
# +
# create the final table with recency, frequency, lifetime
RF = spark.sql("select user, lifetime_indicator, max_days, \
ifnull(freq_times, 0)/30 as frequency, \
(lifetime_indicator*30-max(max_days) \
over(partition by user \
order by lifetime_indicator asc \
range between unbounded preceding and current row)) as recency \
from df_sum2 \
order by user, lifetime_indicator")
RF.createOrReplaceTempView("df_x")
RF.show(60)
# + [markdown] colab_type="text" id="QbGiguZxDZwC"
# ### Q9. Regress recency and frequency on Y and plot the MSE for each lifetime plot
# + colab={} colab_type="code" id="ywqd-cs2D3Re"
from pyspark.ml.regression import LinearRegression
# + colab={} colab_type="code" id="umVTMuaMYjae"
# # join two tables into one table containing frequency, recency, lifetime and number of transactions
df5.createOrReplaceTempView("df5")
RF_final = spark.sql("select RF.user, RF.lifetime_indicator as lifetime, RF.frequency as frequency_within_month, RF.recency as recency_within_month, df5.number_of_transactions_within_12_months from RF inner join df5 on RF.user = df5.user")
# + colab={} colab_type="code" id="I4epa6lojT0C"
RF_final.createOrReplaceTempView("RF_final_2")
t0 = spark.sql("select * from RF_final_2 where lifetime = 0")
t1 = spark.sql("select * from RF_final_2 where lifetime = 1")
t2 = spark.sql("select * from RF_final_2 where lifetime = 2")
t3 = spark.sql("select * from RF_final_2 where lifetime = 3")
t4 = spark.sql("select * from RF_final_2 where lifetime = 4")
t5 = spark.sql("select * from RF_final_2 where lifetime = 5")
t6 = spark.sql("select * from RF_final_2 where lifetime = 6")
t7 = spark.sql("select * from RF_final_2 where lifetime = 7")
t8 = spark.sql("select * from RF_final_2 where lifetime = 8")
t9 = spark.sql("select * from RF_final_2 where lifetime = 9")
t10 = spark.sql("select * from RF_final_2 where lifetime = 10")
t11 = spark.sql("select * from RF_final_2 where lifetime = 11")
t12 = spark.sql("select * from RF_final_2 where lifetime = 12")
# + colab={} colab_type="code" id="R9RM5Fd8lEo0"
# define the linear regression model
def linear_model(data):
from pyspark.ml.regression import LinearRegression
from pyspark.ml.evaluation import RegressionEvaluator
from pyspark.ml.feature import VectorAssembler
vectorAssembler = VectorAssembler(inputCols = ['recency_within_month', 'frequency_within_month','lifetime'], outputCol = 'features')
transformed = vectorAssembler.transform(data)
transformed = transformed.select('features', 'number_of_transactions_within_12_months')
train_df, test_df = transformed.randomSplit([0.7, 0.3], seed = 1)
lr = LinearRegression(featuresCol = 'features', labelCol='number_of_transactions_within_12_months')
lr_model = lr.fit(train_df)
testResults = lr_model.evaluate(test_df)
rmse = testResults.rootMeanSquaredError
mse = rmse**2
return mse
# + colab={"base_uri": "https://localhost:8080/", "height": 307} colab_type="code" id="y792C6zfg3D3" outputId="53e9f314-a1ff-4ea3-f909-0006cd605ee4"
mse_t0 = linear_model(t0)
# -
mse_t0
# + colab={} colab_type="code" id="INtbZMcIv3L0"
mse_t1 = linear_model(t1)
# -
mse_t1
# + colab={} colab_type="code" id="nBul4BXYv3dp"
mse_t2 = linear_model(t2)
# -
mse_t2
# + colab={} colab_type="code" id="3VhUf1kGv3nB"
mse_t3 = linear_model(t3)
# -
mse_t3
# + colab={} colab_type="code" id="uesIZtHTv3rN"
mse_t4 = linear_model(t4)
# -
mse_t4
# + colab={} colab_type="code" id="FGRNPfd-v3v8"
mse_t5 = linear_model(t5)
# -
mse_t5
# + colab={} colab_type="code" id="BkmtICZMv3yz"
mse_t6 = linear_model(t6)
# -
mse_t6
# + colab={} colab_type="code" id="VvpFANKNv31m"
mse_t7 = linear_model(t7)
# -
mse_t7
# + colab={} colab_type="code" id="s58kNs_ev34O"
mse_t8 = linear_model(t8)
# -
mse_t8
# + colab={} colab_type="code" id="BSYzkzuav37H"
mse_t9 = linear_model(t9)
# -
mse_t9
# + colab={} colab_type="code" id="ZNmFyPQYv39_"
mse_t10 = linear_model(t10)
# -
mse_t10
# + colab={} colab_type="code" id="eChQ3OxHv4EY"
mse_t11 = linear_model(t11)
# -
mse_t11
mse_t12 = linear_model(t12)
mse_t12
# + colab={} colab_type="code" id="Dgs1smo5NJsc"
mse = [mse_t0, mse_t1, mse_t2, mse_t3, mse_t4, mse_t5, mse_t6, mse_t7, mse_t8, mse_t9, mse_t10,mse_t11,mse_t12]
# + colab={} colab_type="code" id="aNet94EZNVFq"
import matplotlib.pyplot as plt
# + colab={"base_uri": "https://localhost:8080/", "height": 281} colab_type="code" id="-5OC_eamWZvG" outputId="2c5600cf-d0f7-4e0f-c8ad-4941e916b9e9"
# plot the MSE plot
plt.plot(range(0,13), mse, marker = 'X', c = 'g')
plt.title('MSE Across User Lifetime(0-12)')
plt.xticks(range(0,13),['t0','t1','t2','t3','t4','t5','t6','t7','t8','t9','t10','t11','t12'])
plt.show()
# + [markdown] colab={} colab_type="code" id="FTN41oCg-UGC"
# ### Q10 For each user’s lifetime point, regress recency, frequency AND her spending behavior profile on Y. Plot the MSE for each lifetime point like above. Did you get any improvement?
# + colab={} colab_type="code" id="CftAE8FJ-UKO"
# join the Recency-Frequency table with the spending behavior profile table
combined1 = spark.sql("""select *
from dynamic_profile_clean inner join RF_final on dynamic_profile_clean.User=RF_final.user and dynamic_profile_clean.month=RF_final.lifetime""")
# + colab={} colab_type="code" id="QoefxKmK-UPa"
combined1.show(3)
# + colab={} colab_type="code" id="IjCaPC34-UT1"
from pyspark.ml.feature import VectorAssembler
vectorAssembler = VectorAssembler(inputCols = ['recency_within_month', 'frequency_within_month','activity','cash','event','food','illegal','people','transportation','travel','utility'], outputCol = 'features')
combined1 = vectorAssembler.transform(combined1)
combined1 = combined1.select(['features', 'number_of_transactions_within_12_months','lifetime'])
# + colab={} colab_type="code" id="9fcuw5r7-UWu"
combined1.createOrReplaceTempView("combined1")
t0 = spark.sql("select features, number_of_transactions_within_12_months from combined1 where lifetime = 0")
t1 = spark.sql("select features, number_of_transactions_within_12_months from combined1 where lifetime = 1")
t2 = spark.sql("select features, number_of_transactions_within_12_months from combined1 where lifetime = 2")
t3 = spark.sql("select features, number_of_transactions_within_12_months from combined1 where lifetime = 3")
t4 = spark.sql("select features, number_of_transactions_within_12_months from combined1 where lifetime = 4")
t5 = spark.sql("select features, number_of_transactions_within_12_months from combined1 where lifetime = 5")
t6 = spark.sql("select features, number_of_transactions_within_12_months from combined1 where lifetime = 6")
t7 = spark.sql("select features, number_of_transactions_within_12_months from combined1 where lifetime = 7")
t8 = spark.sql("select features, number_of_transactions_within_12_months from combined1 where lifetime = 8")
t9 = spark.sql("select features, number_of_transactions_within_12_months from combined1 where lifetime = 9")
t10 = spark.sql("select features, number_of_transactions_within_12_months from combined1 where lifetime = 10")
t11 = spark.sql("select features, number_of_transactions_within_12_months from combined1 where lifetime = 11")
# + colab={} colab_type="code" id="e_45_v7R-USC"
mse_t0 = linear_model(t0)
mse_t1 = linear_model(t1)
mse_t2 = linear_model(t2)
mse_t3 = linear_model(t3)
mse_t4 = linear_model(t4)
mse_t5 = linear_model(t5)
mse_t6 = linear_model(t6)
mse_t7 = linear_model(t7)
mse_t8 = linear_model(t8)
mse_t9 = linear_model(t9)
mse_t10 = linear_model(t10)
mse_t11 = linear_model(t11)
# + colab={} colab_type="code" id="kYXOLAUE-UNA"
mse = [mse_t0, mse_t1, mse_t2, mse_t3, mse_t4, mse_t5, mse_t6, mse_t7, mse_t8, mse_t9, mse_t10,mse_t11]
# + colab={} colab_type="code" id="UrG1YFG--UIi"
plt.plot(range(0,12), mse, marker = 'X', c = 'g')
plt.title('MSE Across User Lifetime(0-11)')
plt.xticks(range(0,12),['t0','t1','t2','t3','t4','t5','t6','t7','t8','t9','t10','t11'])
plt.show()
# -
# <img style="float: left;" src="Q10.png" width="40%">
# + [markdown] colab={} colab_type="code" id="hVr8XgBh-UED"
# ### Q11 For each user’s lifetime point, regress her social network metrics on Y. Plot the MSE for each lifetime point like above. What do you observe? How do social network metrics compare with the RF framework? What are the most informative predictors?
# + colab={} colab_type="code" id="SlOf_4wk-UB7"
# join the Recency-Frequency table with the social network metric table
combined2 = spark.sql("""select *
from network_analytic inner join RF_final on network_analytic.user1=RF_final.user and network_analytic.month=RF_final.lifetime""")
# + colab={} colab_type="code" id="YvQfONnJ-T9E"
combined2.show(3)
# -
from pyspark.ml.feature import VectorAssembler
vectorAssembler = VectorAssembler(inputCols = ['friend_num','fof_num','cluster_coef'], outputCol = 'features')
combined2 = vectorAssembler.transform(combined2)
combined2 = combined2.select(['features', 'number_of_transactions_within_12_months','lifetime'])
combined2.show(20)
combined2.createOrReplaceTempView("combined2")
t0 = spark.sql("select features, number_of_transactions_within_12_months from combined2 where lifetime = 0")
t1 = spark.sql("select features, number_of_transactions_within_12_months from combined2 where lifetime = 1")
t2 = spark.sql("select features, number_of_transactions_within_12_months from combined2 where lifetime = 2")
t3 = spark.sql("select features, number_of_transactions_within_12_months from combined2 where lifetime = 3")
t4 = spark.sql("select features, number_of_transactions_within_12_months from combined2 where lifetime = 4")
t5 = spark.sql("select features, number_of_transactions_within_12_months from combined2 where lifetime = 5")
t6 = spark.sql("select features, number_of_transactions_within_12_months from combined2 where lifetime = 6")
t7 = spark.sql("select features, number_of_transactions_within_12_months from combined2 where lifetime = 7")
t8 = spark.sql("select features, number_of_transactions_within_12_months from combined2 where lifetime = 8")
t9 = spark.sql("select features, number_of_transactions_within_12_months from combined2 where lifetime = 9")
t10 = spark.sql("select features, number_of_transactions_within_12_months from combined2 where lifetime = 10")
t11 = spark.sql("select features, number_of_transactions_within_12_months from combined2 where lifetime = 11")
# +
mse = []
for model in [t0,t1,t2,t3,t4,t5,t6,t7,t8,t9,t10,t11]:
mse.append(linear_model(model))
# -
plt.plot(range(0,12), mse, marker = 'X', c = 'g')
plt.title('MSE Across User Lifetime(0-11)')
plt.xticks(range(0,12),['t0','t1','t2','t3','t4','t5','t6','t7','t8','t9','t10','t11'])
plt.show()
# + [markdown] colab={} colab_type="code" id="tFdih_xd-T4H"
# ### Q12 For each user’s lifetime point, regress her social network metrics and the spending behavior of her social network on Y. Plot the MSE for each lifetime point like above. Does the spending behavior of her social network add any predictive benefit compared to Q10?
# +
#calcualte dynamic social network spending behavior
combined3 = spark.sql("""select user1, netbehave_dynamic.month,mean(number_of_transactions_within_12_months) number_of_transactions_within_12_months, mean(friend_num) friend_num, mean(fof_num) fof_num, mean(cluster_coef) cluster_coef, avg(activity) activity, avg(cash) cash, avg(event) event, avg(food) food, avg(illegal) as illegal,
avg(people) people, avg(transportation) transportation, avg(travel) travel, avg(utility) utility
from netbehave_dynamic inner join dynamic_profile_clean on netbehave_dynamic.user2=dynamic_profile_clean.User and netbehave_dynamic.month=dynamic_profile_clean.month
group by netbehave_dynamic.month and netbehave_dynamic.user1""")
# -
from pyspark.ml.feature import VectorAssembler
vectorAssembler = VectorAssembler(inputCols = ['friend_num','fof_num','cluster_coef','activity','cash','event','food','illegal','people','transportation','travel','utility'], outputCol = 'features')
combined3 = vectorAssembler.transform(combined3)
combined3 = combined2.select(['features', 'number_of_transactions_within_12_months','month'])
combined3.createOrReplaceTempView("combined3")
t0 = spark.sql("select features, number_of_transactions_within_12_months from combined3 where month = 0")
t1 = spark.sql("select features, number_of_transactions_within_12_months from combined3 where month = 1")
t2 = spark.sql("select features, number_of_transactions_within_12_months from combined3 where month = 2")
t3 = spark.sql("select features, number_of_transactions_within_12_months from combined3 where month = 3")
t4 = spark.sql("select features, number_of_transactions_within_12_months from combined3 where month = 4")
t5 = spark.sql("select features, number_of_transactions_within_12_months from combined3 where month = 5")
t6 = spark.sql("select features, number_of_transactions_within_12_months from combined3 where month = 6")
t7 = spark.sql("select features, number_of_transactions_within_12_months from combined3 where month = 7")
t8 = spark.sql("select features, number_of_transactions_within_12_months from combined3 where month = 8")
t9 = spark.sql("select features, number_of_transactions_within_12_months from combined3 where month = 9")
t10 = spark.sql("select features, number_of_transactions_within_12_months from combined3 where month = 10")
t11 = spark.sql("select features, number_of_transactions_within_12_months from combined3 where month = 11")
# +
mse = []
for model in [t0,t1,t2,t3,t4,t5,t6,t7,t8,t9,t10,t11]:
mse.append(linear_model(model))
plt.plot(range(0,12), mse, marker = 'X', c = 'g')
plt.title('MSE Across User Lifetime(0-11)')
plt.xticks(range(0,12),['t0','t1','t2','t3','t4','t5','t6','t7','t8','t9','t10','t11'])
plt.show()
| 117,206 |
/using-watson-machine-learning.ipynb | 9d3e3708efbb02e3dea7fa5233decace25527c46 | [] | no_license | akhywali/digitalentAI2019 | https://github.com/akhywali/digitalentAI2019 | 1 | 1 | null | null | null | null | Jupyter Notebook | false | false | .py | 778,136 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# <img src="https://i.imgur.com/hZgPddE.jpg" alt="IBMDBG"> <br>
# # Part 2: Introduction
#
# In this Jupyter Notebook you'll learn step-by-step how to use the Watson Machine Learning API that was automatically generated when the previously created WS Modeler flow was deployed. You will also learn how to download files from IBM Cloud Object Storage and generate interactive visualizations using `Bokeh`.
#
# This Notebook is part of the series on stock market forecasting.
# <hr>
# # Table of Contents
#
# #### 1. Downloading Files from IBM COS
# * 1.1: IBM COS Credentials Setup
# * 1.2: Using the ibm_boto3 Package
#
# #### 2. Reading CSV Files as Pandas Dataframes
# * 2.1: Reading the `AAPL_Test` data
# * 2.2: Reading the `AAPL_Train` data
#
# #### 3. Using the Watson Machine Learning API
# * 3.1: Preparing Input Data
# * 3.2: Setting up WML Credentials
# * 3.3: Making an API Call to WML
# * 3.4: Parsing the results
#
# #### 4. Visualizing the Results
#
# * 4.1: Plotting the Modeler Flow Forecasts
# * 4.2: Validating Modeler Flow Forecasts with Observed Data
# * 4.3: Interacting with Complete Historic and Forecasted Data
# <hr>
# # 1: Downloading Files from IBM COS
# This section objective is to extract files previously stored at IBM Cloud Object Storage:
#
# * The AAPL stocks historical data stored at the `AAPL.csv` file.
# * The Modeler Flow Time Series Model results exported as `AAPL_1-Year_Future_Data.csv` file.
# We will use the `ibm_boto3` library to communicate with IBM Cloud Object Storage.
# +
from ibm_botocore.client import Config
import ibm_boto3
print('Packages imported.')
# -
# ### 1.1: IBM COS Credentials Setup
# Configure your IBM Cloud Object Storage credentials in the cell below.
#
# These credentials can be viewed on the service page instantiated in the IBM Cloud Web page.
# Paste here your IBM COS credentials
cos_credentials = {
'IAM_SERVICE_ID': '',
'IBM_API_KEY_ID': '',
'ENDPOINT': '',
'IBM_AUTH_ENDPOINT': '',
'BUCKET': '',
}
# ### 1.2: Using the ibm_boto3 Package
# Next, we define a function to authenticate with IBM COS and download a defined file.
def download_file_from_cos(credentials, save_file_locally_as, target_file_name):
""" Download a file from IBM COS """
# Configure IBM COS API credentials
cos = ibm_boto3.client(service_name='s3',
ibm_api_key_id=credentials['IBM_API_KEY_ID'],
ibm_service_instance_id=credentials['IAM_SERVICE_ID'],
ibm_auth_endpoint=credentials['IBM_AUTH_ENDPOINT'],
config=Config(signature_version='oauth'),
endpoint_url=credentials['ENDPOINT'])
# Try to download the file
try:
res=cos.download_file(Bucket=credentials['BUCKET'], Key=target_file_name, Filename=save_file_locally_as)
except Exception as e:
print(Exception, e)
else:
print("'{}' file downloaded.".format(target_file_name))
# We use the previously defined function to download the `AAPL.csv_shaped.csv` file.
download_file_from_cos(cos_credentials, 'AAPL_Train.csv', 'AAPL_Train.csv')
download_file_from_cos(cos_credentials, 'AAPL_Test.csv', 'AAPL_Test.csv')
# <hr>
# # 2: Reading CSV Files as Pandas Dataframes
# To generate an interactive graph using the `bokeh` library, we first need to format the data into a panda dataframe.
# +
import pandas as pd
import numpy as np
import os
import dateutil
print('Packages imported.')
# -
# ### 2.1: Reading the `AAPL_Test` data
# Loading the CSV file into a pandas dataframe, with the correct datatypes for each column
dateparse = lambda dates: pd.datetime.strftime(dateutil.parser.parse(dates), '%Y-%m-%d')
df_test = pd.read_csv('AAPL_Test.csv', parse_dates=['Date'], date_parser=dateparse)
print(df_test.info())
df_test = df_test[['Date','Open','High','Low','Close']]
df_test.tail()
# ### 2.2: Reading the `AAPL_Train` data
dateparse = lambda dates: pd.datetime.strftime(dateutil.parser.parse(dates), '%Y-%m-%d')
df_train = pd.read_csv('AAPL_Train.csv', parse_dates=['Date'], date_parser=dateparse)
print(df_train.info())
df_train = df_train[['Date','Open','High','Low','Close']]
df_train.head()
# The `AAPL.csv` file contains the historical data for the Apple Inc. stocks.
#
# The data read as a Pandas dataframe will only be used in Section 4 of this Notebook, for visualization purposes.
# <hr>
# # 3: Using the Watson Machine Learning API
# Previously, we trained a time series forecaster in Watson Modeler Flow and later deployed this forecaster in a Watson Machine Learning service instance.
#
# Now, in this section, we will use the WML API to send new input data to our time series forecaster.
# ### 3.1: Preparing Input Data
# As it can be noted in the cell below, the input data must be a `dict` type and the values must be in the same format as the input CSV file used as data source in Modeler flow.
#
# <img src="https://i.imgur.com/WND3Pqg.png" alt="EX1"> <br>
# The `payload_scoring` dict will contain only some points of data for demonstration.
#
# Remember that the `WIKI/TABLE` Quandl database only goes until 27-March-2018.
#
# The `COLUMN2`, `COLUMN3`, `COLUMN4`, `COLUMN5`, and `COLUMN6` fields are the `DATE`, `OPEN`, `CLOSE`, `HIGH`, `LOW` labels.
# NOTE: manually define and pass the array(s) of values to be scored in the next line
payload_scoring = {"fields": ["COLUMN2", "COLUMN3", "COLUMN4", "COLUMN5", "COLUMN6"],
"values": [['2017-03-28', 140.91, 144.04, 140.62, 143.80]]}
# ### 3.2: Setting up WML Credentials
# Retrieve your `wml_service_credentials_username`, `wml_service_credentials_password`, and `wml_service_credentials_url` from the service credentials associated with your IBM Cloud Watson Machine Learning Service instance.
#
# This can be done accessing the service instance in the IBM Cloud web portal.
# +
wml_credentials = {
"password": "",
"url": "",
"username": ""
}
deployment_endpoint = ""
# -
# ### 3.3: Making an API Call to WML
# The next code cell uses `urllib3` and `requests` to communicate with the WML API. The `payload_scoring` dict is used as input.
# +
import urllib3, requests, json
headers = urllib3.util.make_headers(basic_auth='{username}:{password}'.format(username=wml_credentials['username'], password=wml_credentials['password']))
url = '{}/v3/identity/token'.format(wml_credentials['url'])
response = requests.get(url, headers=headers)
mltoken = json.loads(response.text).get('token')
header = {'Content-Type': 'application/json', 'Authorization': 'Bearer ' + mltoken}
response_scoring = requests.post(deployment_endpoint, json=payload_scoring, headers=header)
print("Scoring finished")
print("Response type: {}".format(type(json.loads(response_scoring.text))))
# -
# In the next cell we can visualize the WML response, that is also of type `dict`.
data = json.loads(response_scoring.text)
print(data)
# Before using `Bokeh` to interact with the data, we need to parse it in a Pandas dataframe.
# ### 3.4: Parsing the WML Results
# +
from datetime import datetime
def parse(dic):
for k, v in dic.items():
if isinstance(v, dict):
for p in parse(v):
yield [k] + p
else:
yield [k, v]
lst = list(parse(data))
columns = lst[0][1]
values = lst[1][1]
def parse(values):
for k in values:
string_lst = k[0].split(" ")
k[0] = datetime.strptime(string_lst[0], '%Y-%m-%d')
parse(values)
# -
# The code cell above transformed the `dict` response into two lists: the labels (columns) and rows (values).
print(values)
# Next we just create a new Pandas dataframe with the future data for Apple Inc. stocks, retrieved from WML using the API.
ndf = pd.DataFrame.from_records(values, columns=columns)
print(ndf.info())
ndf.tail()
# <hr>
# # 4: Validating and Visualizing the Results
# +
from bokeh.plotting import figure, output_file, show
from bokeh.models import ColumnDataSource
from bokeh.embed import components
from bokeh.io import output_notebook
print('Packages imported.')
# -
# Load bokeh
output_notebook()
# ### 4.1: Plotting the Modeler Flow Forecasts
# +
# Figure
p = figure(plot_width=1200, plot_height=550, title='Historic and Predicted Stock Value Data', x_axis_type="datetime")
# Plot Lines
p.line(ndf.Date, ndf['$TS-Close'], line_width=3, line_color="#ff6699", legend='Modeled Close Value')
p.line(ndf.Date, ndf['$TS-Open'], line_width=3, line_color="#0099ff", legend='Modeled Open Value')
p.line(ndf.Date, ndf['$TSLCI-Close'], line_width=0.5, line_color="#ff6699", legend='Modeled Close Value Bounds')
p.line(ndf.Date, ndf['$TSUCI-Close'], line_width=0.5, line_color="#ff6699", legend='Modeled Close Value Bounds')
p.line(ndf.Date, ndf['$TSLCI-Open'], line_width=0.5, line_color="#0099ff", legend='Modeled Open Value Bounds')
p.line(ndf.Date, ndf['$TSUCI-Open'], line_width=0.5, line_color="#0099ff", legend='Modeled Open Value Bounds')
# Axis and Labels
p.legend.orientation = "vertical"
p.legend.location = "top_left"
p.xaxis.axis_label = "Date"
p.xaxis.axis_label_text_font_style = 'bold'
p.xaxis.axis_label_text_font_size = '16pt'
p.xaxis.major_label_text_font_size = '14pt'
p.yaxis.axis_label = "Value ($ USD)"
p.yaxis.axis_label_text_font_style = 'bold'
p.yaxis.axis_label_text_font_size = '16pt'
p.yaxis.major_label_text_font_size = '12pt'
# -
show(p)
# ### 4.2: Validating Modeler Flow Forecasts with Observed Data
# +
# Figure
p = figure(plot_width=1200, plot_height=550, title='Historic and Predicted Stock Value Data', x_axis_type="datetime")
# Plot Lines
p.line(ndf.Date, ndf['$TS-Close'], line_width=3, line_color="#ff6699", legend='Forecasted Close Value')
p.line(ndf.Date, ndf['$TS-Open'], line_width=3, line_color="#0099ff", legend='Forecasted Open Value')
p.line(df_test[df_test['Date'] > datetime(2015,1,1)].Date, df_test[df_test['Date'] > datetime(2015,1,1)].Close, line_width=0.5, line_color="#ff6699", legend='Historic Close Data (Test Sample)')
p.line(df_test[df_test['Date'] > datetime(2015,1,1)].Date, df_test[df_test['Date'] > datetime(2015,1,1)].Open, line_width=0.5, line_color="#0099ff", legend='Historic Open Data (Test Sample)')
# Axis and Labels
p.legend.orientation = "vertical"
p.legend.location = "top_left"
p.xaxis.axis_label = "Date"
p.xaxis.axis_label_text_font_style = 'bold'
p.xaxis.axis_label_text_font_size = '16pt'
p.xaxis.major_label_text_font_size = '14pt'
p.yaxis.axis_label = "Value ($ USD)"
p.yaxis.axis_label_text_font_style = 'bold'
p.yaxis.axis_label_text_font_size = '16pt'
p.yaxis.major_label_text_font_size = '12pt'
# -
show(p)
# +
ndf_filtered = ndf.drop(['Close', 'Open', '$TSResidual-Open', '$TSResidual-Close'], axis=1)
result = pd.concat([ndf_filtered, df_test], axis=1).dropna()
result.tail()
# -
# In the next cell, simple mean errors are calculated (percentual and absolute):
# +
open_abs_errors = []
close_abs_errors = []
open_pct_errors = []
close_pct_errors = []
for index, row in result.iterrows():
open_abs_errors.append(abs(row['Open']-row['$TS-Open']))
close_abs_errors.append(abs(row['Close']-row['$TS-Close']))
open_pct_errors.append((abs(row['Open']-row['$TS-Open']))/row['Open'])
close_pct_errors.append((abs(row['Close']-row['$TS-Close']))/row['Close'])
mean_open_error = sum(open_abs_errors) / len(open_abs_errors)
mean_close_error = sum(close_abs_errors) / len(close_abs_errors)
mean_open_pct_error = sum(open_pct_errors) / len(open_pct_errors)
mean_close_pct_error = sum(close_pct_errors) / len(close_pct_errors)
print('Mean Errors in 1-Year Future Prediction:')
print('Analyzed Stock: AAPL (Apple Inc.)')
print('----------------------------------------')
print('Mean Open Value Error (USD): {} $'.format(round(mean_open_error, 3)))
print('Mean Close Value Error (USD): {} $'.format(round(mean_close_error, 3)))
print('Mean Open Value Error: {}%'.format(round(mean_open_pct_error*100, 3)))
print('Mean Close Value Error: {}%'.format(round(mean_close_pct_error*100, 3)))
# -
# ### 4.3: Interacting with Complete Historic and Forecasted Data
# +
# Figure
p = figure(plot_width=1200, plot_height=550, title='Historic and Predicted Stock Value Data', x_axis_type="datetime")
# Plot Lines
p.line(ndf.Date, ndf['$TSLCI-Close'], line_width=0.5, line_color="#ff6699", legend='Modeled Close Value Bounds')
p.line(ndf.Date, ndf['$TSUCI-Close'], line_width=0.5, line_color="#ff6699", legend='Modeled Close Value Bounds')
p.line(ndf.Date, ndf['$TSLCI-Open'], line_width=0.5, line_color="#0099ff", legend='Modeled Open Value Bounds')
p.line(ndf.Date, ndf['$TSUCI-Open'], line_width=0.5, line_color="#0099ff", legend='Modeled Open Value Bounds')
p.line(df_train.Date, df_train['Open'], line_width=0.5, line_color="#0099ff", legend='Historic Open Data (Train Sample)')
p.line(df_train.Date, df_train['Close'], line_width=0.5, line_color="#ff6699", legend='Historic Close Data (Train Sample)')
p.line(ndf.Date, ndf['$TS-Close'], line_width=3, line_color="#ff6699", legend='Forecasted Close Value')
p.line(ndf.Date, ndf['$TS-Open'], line_width=3, line_color="#0099ff", legend='Forecasted Open Value')
p.line(df_test[df_test['Date'] > datetime(2015,1,1)].Date, df_test[df_test['Date'] > datetime(2015,1,1)].Close, line_width=0.5, line_color="#ff6699", legend='Historic Close Data (Test Sample)')
p.line(df_test[df_test['Date'] > datetime(2015,1,1)].Date, df_test[df_test['Date'] > datetime(2015,1,1)].Open, line_width=0.5, line_color="#0099ff", legend='Historic Open Data (Test Sample)')
# Axis and Labels
p.legend.orientation = "vertical"
p.legend.location = "top_left"
p.xaxis.axis_label = "Date"
p.xaxis.axis_label_text_font_style = 'bold'
p.xaxis.axis_label_text_font_size = '16pt'
p.xaxis.major_label_text_font_size = '14pt'
p.yaxis.axis_label = "Value ($ USD)"
p.yaxis.axis_label_text_font_style = 'bold'
p.yaxis.axis_label_text_font_size = '16pt'
p.yaxis.major_label_text_font_size = '12pt'
# -
show(p)
# <hr>
#
# This notebook and its source code is made available under the terms of the <a href = "https://github.com/vanderleipf/ibmdegla-ws-projects/blob/master/LICENSE">MIT License</a>.
#
# <hr>
# ### Thank you for completing this journey!
#
# Notebook created by: <a href = "https://www.linkedin.com/in/vanderleimpf87719/">Vanderlei Pereira</a>
None
# case 2
elif self.current_node.left != None and self.current_node.right == None :
if value < self.parent.value :
self.parent.left = self.current_node.left
else :
self.parent.right = self.current_node.left
elif self.current_node.left == None and self.current_node.right != None :
if value < self.parent.value :
self.parent.left = self.current_node.right
else :
self.parent.right = self.current_node.right
# case 3
elif self.current_node.left != None and self.current_node.right != None :
# case 3-1
if value < self.parent.value :
self.change_node = self.current_node.right
self.change_node_parent = self.current_node.right
while self.change_node.left != None :
self.change_node_parent = self.change_node
self.change_node = self.change_node.left
if self.change_node.right != None :
self.change_node_parent.left = self.change_node.right
else :
self.change_node_parent.left = None
self.parent.left = self.change_node
self.change_node.right = self.current_node.right
self.change_node.left = self.change_node.left
# case 3-2
else :
self.chagne_node = self.current_node.right
self.change_node_parent = self.current_node.right
while self.change_node.left != None :
self.change_node_parent = self.change_node
self.change_node = self.change_node.left
if self.change_node.right != None :
self.change_node_parent.left = self.change_node.right
else :
self.change_node_parent.left = None
self.parent.right = self.change_node
self.change_node.right = self.current_node.right
self.change_node.left = self.current_node.left
return True
# -
# 참고: http://ejklike.github.io/2018/01/09/traversing-a-binary-tree-1.html
# #### 7-5-5-6. 파이썬 전체 코드 테스트
# - random 라이브러리 활용
# - random.randint(첫번째 숫자, 마지막 숫자): 첫번째 숫자부터 마지막 숫자 사이에 있는 숫자를 랜덤하게 선택해서 리턴
# - 예: random.randint(0, 99): 0에서 99까지 숫자중 특정 숫자를 랜덤하게 선택해서 리턴해줌
# +
# 0 ~ 999 숫자 중에서 임의로 100개를 추출해서, 이진 탐색 트리에 입력, 검색, 삭제
import random
# 0 ~ 999 중, 100 개의 숫자 랜덤 선택
bst_nums = set()
while len(bst_nums) != 100 :
bst_nums.add(random.randint(0, 999))
# print(bst_nums)
# 선택된 100개의 숫자를 이진 탐색 트리에 입력, 임의로 루트노드는 500을 넣기로 함
head = Node(500)
binary_tree = NodeMgmt(head)
for num in bst_nums :
binary_tree.insert(num)
# 입력한 100개의 숫자 검색 (검색 기능 확인)
for num in bst_nums :
if binary_tree.search(num) == False :
print("search failed", num)
# 입력한 100개의 숫자 중 10개의 숫자를 랜덤 선택
delete_nums = set()
bst_nums = list(bst_nums)
while len(delete_nums) != 10 :
delete_nums.add(bst_nums[random.randint(0, 99)])
# 선택한 10개의 숫자를 삭제 (삭제 기능 확인)
for del_num in delete_nums :
if binary_tree.delete(del_num) == False :
print("delete failed", del_num)
# -
# ### 7-6. 이진 탐색 트리의 시간 복잡도와 단점
# #### 7-6-1. 시간 복잡도 (탐색시)
# - depth (트리의 높이) 를 h라고 표기한다면, O(h)
# - n개의 노드를 가진다면, $h = log_2{n} $ 에 가까우므로, 시간 복잡도는 $ O(log{n}) $
# - 참고: 빅오 표기법에서 $log{n}$ 에서의 log의 밑은 10이 아니라, 2
# - 한번 실행시마다, 50%의 실행할 수도 있는 명령을 제거한다는 의미. 즉 50%의 실행시간을 단축시킬 수 있다는 것을 의미
# <br>
# <img src="https://www.mathwarehouse.com/programming/images/binary-search-tree/binary-search-tree-sorted-array-animation.gif" />
# <br>
# (출처: https://www.mathwarehouse.com/programming/gifs/binary-search-tree.php#binary-search-tree-insertion-node)
# #### 7-6-2. 이진 탐색 트리 단점
# - 평균 시간 복잡도는 $ O(log{n}) $ 이지만,
# - 이는 트리가 균형잡혀 있을 때의 평균 시간복잡도이며,
# - 다음 예와 같이 구성되어 있을 경우, 최악의 경우는 링크드 리스트등과 동일한 성능을 보여줌 ( $O(n)$ )
# <br>
# <img src="http://www.fun-coding.org/00_Images/worstcase_bst.png" width="300" />
# <br>
# - Miscellaneous feature not covered in other categories
#
# Elev Elevator
# Gar2 2nd Garage (if not described in garage section)
# Othr Other
# Shed Shed (over 100 SF)
# TenC Tennis Court
# NA None
# +
#categorical(train_df, "MiscFeature")
# -
# # Variable 76: MiscVal
# - $Value of miscellaneous featuredv
continuous(train_df, "MiscVal")
# # Variable 77: MoSold
# - Month Sold (MM)
categorical(train_df, "MoSold")
# # Variable 78: YrSold
# - Year Sold (YYYY)
categorical(train_df, "YrSold")
# # Variable 79: SaleType
# - Type of sale
#
# WD Warranty Deed - Conventional
# CWD Warranty Deed - Cash
# VWD Warranty Deed - VA Loan
# New Home just constructed and sold
# COD Court Officer Deed/Estate
# Con Contract 15% Down payment regular terms
# ConLw Contract Low Down payment and low interest
# ConLI Contract Low Interest
# ConLD Contract Low Down
# Oth Other
categorical(train_df, "SaleType")
# # Variable 80: SaleCondition
# - SaleCondition: Condition of sale
#
# Normal Normal Sale
# Abnorml Abnormal Sale - trade, foreclosure, short sale
# AdjLand Adjoining Land Purchase
# Alloca Allocation - two linked properties with separate deeds, typically condo with a garage unit
# Family Sale between family members
# Partial Home was not completed when last assessed (associated with New Homes)
categorical(train_df, "SaleCondition")
# # Variable 81: SalePrice
continuous(train_df, "SalePrice")
# # Variable 85: LogSalePrice
continuous(train_df, "LogSalePrice")
| 20,218 |
/数据结构与算法之美/.ipynb_checkpoints/Chapter.15 二分查找-checkpoint.ipynb | 5f426fa7624d0c0ba5c0b364b7c9de695d4a20fe | [] | no_license | marslwxc/read-project | https://github.com/marslwxc/read-project | 0 | 0 | null | null | null | null | Jupyter Notebook | false | false | .py | 6,769 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# **二分查找针对的是一个有序的数据集合,查找思想有点类似分治思想。每次都通过跟区间的中间元素对比,将带查找的区间缩小为之前的一半,直到找到要查找的元素,或者区间被缩小为0**
# 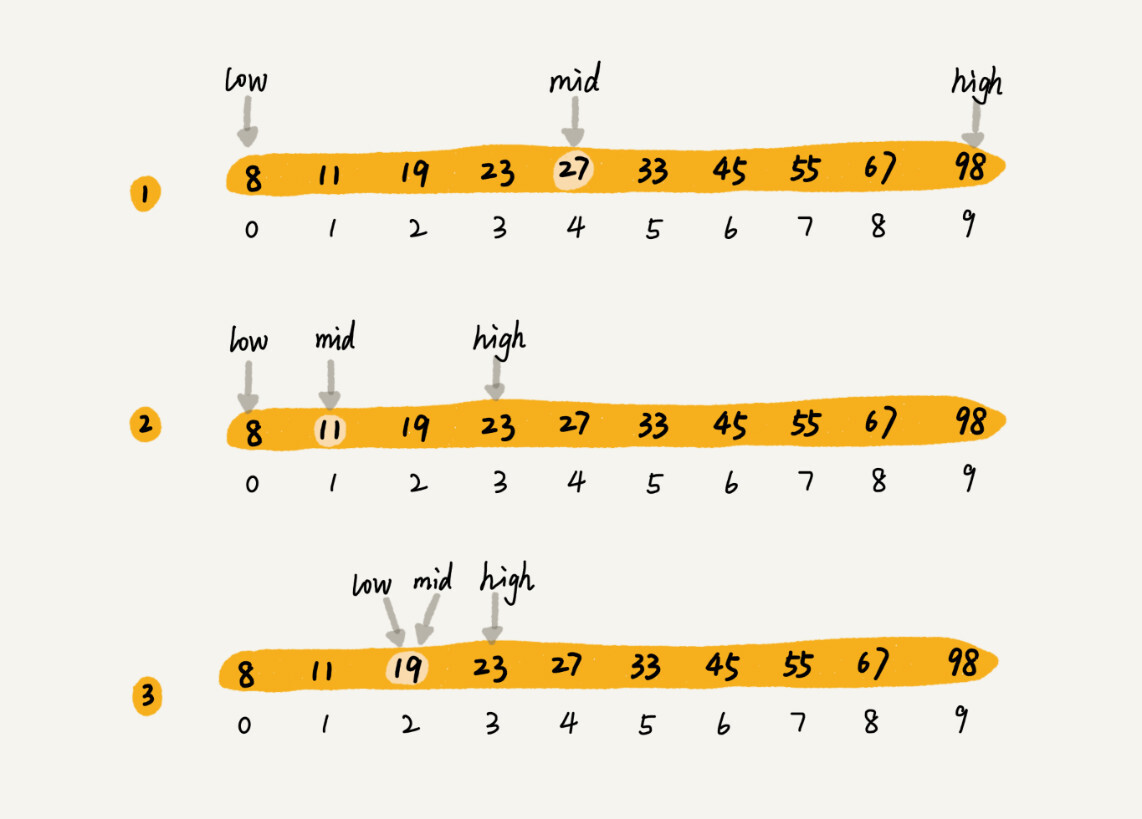
# 1. 二分查找的时间复杂度为O(nlogn)
# 2. 二分查找容易出错的三个地方
# 1. 循环推出条件
# 2. mid的取值
# 3. low和high的更新
def bsearch(a, n):
low, high = 0, len(a)-1
while low<=high:
mid = low + (high - low) // 2
if a[mid] == n:
return mid
if a[mid] < n:
low = mid + 1
else:
high = mid - 1
return -1
# +
def bsearch(nums, target: int):
return bsearch_internally(nums, 0, len(nums)-1, target)
def bsearch_internally(nums, low, high, target):
if low > high:
return -1
mid = low+int((high-low) >> 2)
if nums[mid] == target:
return mid
elif nums[mid] < target:
return bsearch_internally(nums, mid+1, high, target)
else:
return bsearch_internally(nums, low, mid-1, target)
# -
# 二分查找应用场景的局限性
# 1. 二分查找依赖的是顺序表结构,简单点说就是数组
# 2. 二分查找针对的是有序数据
# 3. 数据量太小不合适二分查找
# 4. 数据量太大也不适合二分查找
# #### 变体一:查找第一个值等于给定值的元素
# 有序数据集合中存在重复的数据,我们希望找到第一个值等于给定值的数据
#
# a[mid]跟要查找的value的大小关系有三种情况:大于、小于、等于。
# 1. 对于a[mid]>value的情况,我们需要更新high=mid-1;
# 2. 对于a[mid]<value的情况,我们需要更新low=mid+1。
# 3. 对于a[mid]=value的情况,
# 1. 如果mid等于0,那这个元素已经是数组的第一个元素,那就是我们要找的
# 2. 如果mid不等于0,但a[mid]的前一个元素a[mid-1]不等于value,也说明a[mid]就是我们要找的等一个值等于给定值的元素
# 3. 如果mid的前一个元素也等于value,那说明这个数不是第一个值。那我们就更新high=mid-1
# +
def bsearch_first(a, n):
low, high = 0, len(a)-1
while low<=high:
mid = low + (high - low) // 2
if a[mid] > n:
high = mid - 1
if a[mid] < n:
low = mid + 1
if a[mid] == n:
if mid == 0 or a[mid-1] != n:
return mid
else:
high = mid - 1
return False
a = [1,2,3,4,4,4,5]
n = bsearch_first(a, 4)
print(n)
# -
# #### 变体二:查找最后一个等于给定值的元素
# 1. 省略
# 2. 省略
# 3. a[mid] == value
# 1. 如果a[mid]是数组最后一个元素或者a[mid+1]!=value,这就是我么要的值
# 2. 如果a[mid+1]也等于value,那么low=mid+1
# +
def bsearch_first(a, n):
low, high = 0, len(a)-1
while low<=high:
mid = low + (high - low) // 2
if a[mid] > n:
high = mid - 1
if a[mid] < n:
low = mid + 1
if a[mid] == n:
if mid == 0 or a[mid+1] != n:
return mid
else:
low = mid + 1
return False
a = [1,2,3,4,4,4,5]
n = bsearch_first(a, 4)
print(n)
# -
# #### 变体三:查找第一个大于等于给定值的元素
# 1. 如果a[mid]小于value,那更新low=mid+1
# 2. 如果a[mid]大于等于value
# 1. 如果a[mid]前面已经没有元素,或者前面的值小于value,那么a[mid]就是要找的元素
# 2. 如果a[mid]前面的元素也大于等于value,更新high=mid-1
# +
def bsearch_first(a, n):
low, high = 0, len(a)-1
while low<=high:
mid = low + (high - low) // 2
if a[mid] < n:
low = mid + 1
if a[mid] >= n:
if mid == 0 or a[mid-1] < n:
return mid
else:
high = mid - 1
return False
a = [1,2,3,4,4,4,5]
n = bsearch_first(a, 4)
print(n)
# -
# #### 变体四:查找最后一个小于等于给定值的元素
# 1. 如果a[mid]大于于value,那更新high=mid-1
# 2. 如果a[mid]小于于等于value
# 1. 如果a[mid]后面已经没有元素,或者后面的值大于value,那么a[mid]就是要找的元素
# 2. 如果a[mid]后面的元素也小于等于value,更新low=mid+1
# +
def bsearch_first(a, n):
low, high = 0, len(a)-1
while low<=high:
mid = low + (high - low) // 2
if a[mid] > n:
high = mid - 1
if a[mid] <= n:
if mid == 0 or a[mid+1] > n:
return mid
else:
low = mid + 1
return False
a = [1,2,3,4,4,4,5]
n = bsearch_first(a, 4)
print(n)
# -
| 3,907 |
/수업/Python 프로그래밍 이해(2)_21.07.07/12.파이썬 파일 입출력.ipynb | b908685cd30adae44fcb9dee1fcdfa321d519a1c | [] | no_license | dch9610/Bigdata-_Intern | https://github.com/dch9610/Bigdata-_Intern | 1 | 0 | null | null | null | null | Jupyter Notebook | false | false | .py | 12,930 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# +
# First, check if the DB file for this notebook exists and remove it.
import os
if os.path.exists('example.db'):
os.remove('example.db')
# +
import sqlite3
from sqlalchemy import create_engine
engine = create_engine('sqlite:///example.db', echo=True)
# -
# For example, use a "Declarative Base" object to use an "object-model" approach. We can defined Tables by themselves using SQLAlchemy, but we're not going to do that right now.
from sqlalchemy.ext.declarative import declarative_base
Base = declarative_base()
# This uses the Declarative style to create a "records" object with the fields we want.
# Please note that in the following definition, by keeping only one "detector" and one "enabled" field, we remove the ability for unique indexing on the SeriesNumber and EventNumber fields.
# +
from sqlalchemy import Column, Integer, BigInteger, String, DateTime, Float, Boolean
from sqlalchemy import Sequence
class RecordModel(Base):
__tablename__ = 'records'
recordID = Column(Integer,
Sequence('record_id_seq'),
primary_key=True)
SeriesNumber = Column(BigInteger,
index=True,
#unique=True,
nullable=False)
EventNumber = Column(BigInteger,
index=True,
#unique=True,
nullable=False)
DetNum = Column(Integer)
DetType = Column(Integer)
Enabled = Column(Boolean)
def __repr__(self):
return "<Record(recordID='%s', SeriesNumber='%s', EventNumber=''%s')" % (
self.recordID, self.SeriesNumber, self.EventNumber)
# -
# Using the declarative style automatically creates the indexed "metadata schema" for the database, which will speed up queries and make the database easily sharable.
RecordModel.__table__
# Now let's actually create the SQLite database:
Base.metadata.create_all(engine)
# Let's create some example entries, and insert them in the database.
# +
from sqlalchemy.orm import sessionmaker
# Use a 'factory' for creating a connection to our database.
Session = sessionmaker(bind=engine)
# Create actual session object we will use to interact with DB
session = Session()
# +
# %%time
# Create some example records
import random
import numpy as np
before_commit = []
for i in range(0,8500):
for j in range(0, 35):
_enable = random.choice([True, False])
new_record = RecordModel(SeriesNumber=1005,
EventNumber=500+i,
DetNum=j,
DetType=int(j%10),
Enabled=_enable)
before_commit.append(new_record)
# +
# how many objects?
print(len(before_commit))
# +
# %%time
# Add them to the session and save session to DB, aka "Database Transaction"
session.bulk_save_objects(before_commit)
session.commit()
# -
# What's our resulting filesize? (n.b. Size is in "MebiBytes" [MiB]).
# +
import os
bytecount = os.stat('example.db').st_size
print("DB Size is ", (bytecount * (9.537E-7)), " [MiB]")
# +
# Now I want to form the Pass/Cut True/False array for one particular series+event
target_series = 1005
target_event = (4700)+500
eval_query = session.query(RecordModel).order_by(RecordModel.DetNum).\
filter(RecordModel.SeriesNumber == target_series).\
filter(RecordModel.EventNumber == target_event)
pass_cuts = np.array([record.Enabled for record in eval_query], dtype=bool)
pass_cuts
# +
cuts_dettype = np.array([record.DetType for record in eval_query], dtype=np.int32)
cuts_dettype
# -
# ## do it from scratch
# +
## Set up the basics
from sqlalchemy import create_engine
from sqlalchemy.orm import sessionmaker
new_connection = create_engine('sqlite:///example.db', echo=True)
# Use a 'factory' for creating a connection to our database.
Session = sessionmaker(bind=new_connection)
# Create actual session object we will use to interact with DB
new_session = Session()
## Automap schema from the database itself
from sqlalchemy.ext.automap import automap_base
from sqlalchemy.orm import Session
Base = automap_base()
Base.prepare(new_connection, reflect=True)
RecordModel = Base.classes.records
## And query
import numpy as np
target_series = 1005
target_event = (4700)+500
eval_query = new_session.query(RecordModel).order_by(RecordModel.DetNum).\
filter(RecordModel.SeriesNumber == target_series).\
filter(RecordModel.EventNumber == target_event)
pass_cuts = np.array([record.Enabled for record in eval_query], dtype=bool)
pass_cuts
# -
| 5,001 |
/F-NODE18.ipynb | 6ddc2a2abf65fc9d886d372c79e6d4e6b11a0652 | [] | no_license | Ki8888/first-repository | https://github.com/Ki8888/first-repository | 0 | 0 | null | null | null | null | Jupyter Notebook | false | false | .py | 10,020 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # 파이썬으로 이미지파일 다루기
# ## 디지털 이미지
# * 디지털 화면은 수많은 점들로 이루어져있으며, 색상이 가지는 점 하나를 화소(pixel)이라고 한다.<br> 각 pixel은 R,G,B(Red, Green, Blue) 세개의 단일 색의 강도를 조절하여 색상표현
# * 비트맵(bitmap)방식 : 한점마다 색상별로 8비트를 사용하여 0~255사이 값으로 해당색의 감도 표시
# * 벡터(vector)방식 : 상대적인 점과 선의 위치를 방정식으로써 기록해두었다가, 확대 및 축소에 따라 디지털 화면의 각화소에 어떻게 표현될지 계산 >> 깨짐이 없음(확대, 축소 가능한 글꼴)
# * 색을 구성하는 방식 >>컬러스페이스, 스페이스 구성 단일축(R, G, B) >>채널(chanel)
# jpeg : 근처의 화소를 묶어 비슷한색을 뭉뚱그리는 방식 >>재압축 반복시 색상지저분(디지털풍화)<br>
# PNG : 색상의 손실없이 이미지 압축, 사용색상많아지면 jpeg보다 큰 용량 차지
# ## Pillow 사용법
# * numpy와 결합하여 편리하게 사용가능한 이미지처리 도구
# * 이미지는 배열형태의 데이터 >>>32픽셀, 3색상 >> [32, 32, 3]
# +
# 32 X 32 pixel짜리 검은색 화면
import numpy as np
from PIL import Image
data = np.zeros([32, 32, 3], dtype=np.uint8) #256사이의 값으로 데이터(1pixel) 표현가능하므로 uint8
image = Image.fromarray(data, 'RGB')
image.show()
# +
data[:, :] = [255, 0, 0]
image = Image.fromarray(data, 'RGB')
image.show()
# [[* ][* ][* ]] R (255X255X1)
# [[* ][* ][* ]] G "
# [[* ][* ][* ]] B " >>>255X255X3
# -
# 가로 세로 각 128 픽셀짜리 흰색 이미지 만들기
data1 = np.zeros([128,128,3], dtype=np.uint8)
data1[:,:] = [255, 255, 255]
image1 = Image.fromarray(data1, 'RGB')
image1.show()
# * 연습용 이미지를 열어 width(가로)와 height(세로)를 출력하고 save를 이용하여 jpg포맷으로 저장하기
# +
from PIL import Image
import os
# 연습용파일 경로
image_path = os.getenv('HOME')+'/aiffel/python_image_proc/pillow_practice.png'
# 이미지 열기 (open이용)
img = Image.open(image_path)
img.show()
# width와 height 출력
print(img.width)
print(img.height)
# JPG 파일 형식으로 저장해보기
new_image_path = os.getenv('HOME')+'/aiffel/python_image_proc/pillow_practice.jpg'
img = img.convert('RGB')
img.save(new_image_path)
# -
# * resize() 이용하여 이미지 크기를 100 * 200으로 변경하여 저장
# +
resized_image = img.resize((100, 200))
resized_image.show()
# save
resized_image_path = os.getenv('HOME')+'/aiffel/python_image_proc/pillow_practice_resized.png'
resized_image.save(resized_image_path)
# -
# * crop() 이용하여 눈 부분만 잘라내어 저장하기 (눈부분 box좌표: 300,100,600,400)
# +
box = (300, 100, 600, 400)
region = img.crop(box)
region.show()
cropped_image_path = os.getenv('HOME')+'/aiffel/python_image_proc/pillow_practice_cropped.png'
region.save(cropped_image_path)
# -
# ## Pillow를 활용한 데이터 전처리
# ### CIFAR-100 데이터를 받아 개별 이미지 파일로 추출하기
# * 실습 파일(CIFAR-100 python version) : 32 * 32화소 이미지 100개 클래스당 600장(train:500, test:100) 총 6만장
# +
import os
import pickle
dir_path = os.getenv("HOME")+'/aiffel/python_image_proc/cifar-100-python'
# os.path.join 경로를 병합하여 새 경로 생성
train_file_path = os.path.join(dir_path, 'train') # dir_path/train?
# with문 나올때 close 자동으로 불러줌
# 대부분 인코딩 형태 (읽기-rb, 쓰기-wb, 한작업엔 한개만 하는게 좋음)
with open(train_file_path, 'rb') as f:
train = pickle.load(f, encoding='bytes')
type(train)
# -
train.keys()
type(train[b'filenames'])
train[b'filenames'][0:5]
train[b'data'][0:5]
train[b'data'][0].shape
# * 3072 = 3채널 * 1024(32*32)<br>
# * numpy배열을 reshape하면 이미지파일 원본 복구 가능
# reshape를 앞에서채우는방식이 아닌 원하는 형태로
image_data = train[b'data'][0].reshape([32,32,3], order='F')
# Pillow를 사용하여 Numpy 배열을 Image객체로 만들어서
image = Image.fromarray(image_data)
image.show()
# * 사이즈가 작지만 정상이미지, but X축, Y축 뒤집어져 나옴
# 축 바꿔주기
# np.swapaxes(0,1)
image_data = image_data.swapaxes(0,1)
image.show()
# * 이미지파일을 실제파일처럼 만들어주는 반복작업을 tqdm이용하여 시각화 가능
# +
import os
import pickle
from PIL import Image
import numpy
from tqdm import tqdm
dir_path = os.getenv('HOME')+'/aiffel/python_image_proc/cifar-100-python'
train_file_path = os.path.join(dir_path, 'train')
# image를 저장할 cifar-100-python의 하위디렉토리(images)를 생성합니다.
# /cifar-100-python/images
images_dir_path = os.path.join(dir_path, 'images')
if not os.path.exists(images_dir_path):
os.mkdir(images_dir_path)
# 32*32 이미지파일 50000개를 생성
with open(train_file_path, 'rb') a f:
train = pickle.load(f, encoding='bytes')
for i in tqdm(range(len(train[b'filenames'])))
# -
| 4,064 |
/exploratoryDataAnalysis/californai housing EDA.ipynb | 6624b82ab7f988586edc3d7a15d72f0b4d8d1b47 | [] | no_license | wolfdale229/100daysofmlcodes | https://github.com/wolfdale229/100daysofmlcodes | 0 | 0 | null | null | null | null | Jupyter Notebook | false | false | .py | 1,534,531 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: 'Python 3.7.4 64-bit (''base'': conda)'
# language: python
# name: python37464bitbaseconda02803511e82243628460d84236bfc9e6
# ---
# # Problem :
#
# #### Preforming exploratory data analysis on the `callifornia housing` data set.
#
# # Aim :
# * Visualizing statistical relationship from features in the californai housing dataset to again insights on what house feature contributes to it's price.
# * Use machine learning to model an application that would be able to predict housing prices when given a set of parameters (based of this data set).
#
# # Data :
# #### The data is gotten from the [kaggle website](https://www.kaggle.com/c/californiahousing)
# %reload_ext autoreload
# %autoreload 2
# %matplotlib inline
# +
import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
sns.set_style('darkgrid')
# -
file = '../data/california/housing.csv'
# !ls {file}
df = pd.read_csv(file, sep=',')
# #### Looking at the first five data samples of the data set
df.head()
# #### Let's look at some statistical attributes of the dataset
df.describe()
df.info()
# #### 1 Before we start visualizing the data, we would like to add some additional features. Looking at the data set we could a two additional columns which would represent ` Number of rooms in each houshold` and `Number of bedrooms in each houshold`.
#
# #### 2 Since countable items do not come in decimal points we would change the data type of the newly created features to type `int`. But before we do that, from the statistical description of the dataset we would notice the `total_bedrooms` count is 20433 from a total count of about 20640 which is about `207` points `missing/not computed` during data collection.
#
df['bedrooms'] = round(df['total_bedrooms'] / df['households'])
df['rooms'] = round(df['total_rooms'] / df['households'])
df['bedrooms'].fillna(np.median, inplace=True, axis=0)
df['total_bedrooms'].fillna(np.median, inplace=True, axis=0)
df
df['total_bedrooms'] = pd.to_numeric(df['total_bedrooms'], errors='coerce')
df['bedrooms'] = pd.to_numeric(df['bedrooms'], errors='coerce')
df.isnull().sum()
# ### Extra feature engineering:
# For convinence the median_house name is changed to price, and the median_income would be up scaled to it original values.
df['price'] = df['median_house_value']
df.drop('median_house_value', axis=1,inplace=True)
df['median_income'] = df['median_income'] * 10000
# # Visualization
df.head()
# #### Let's plot a scatter plot of the `median_income` and the `price`
sns.relplot(x='median_income', y='price', data=df, height=8, )
plt.show()
# #### From the above graph it indicates that families with a `median_income` below the #8000 mark purchase houses with prices lower than $400000.
# #### Let's plot a scatter plot of the `median_income` and the `price` based on `median age`
sns.relplot(x='median_income', y='price', data=df,
hue='housing_median_age', height=8, palette='jet')
plt.show()
# #### The scatter plot shows that age has little influence of income distribution.
# #### Let's check out the wealth and `population` distribution
sns.relplot(x='latitude', y='longitude', data=df,
hue='population', palette='jet', height=8)
plt.show()
# #### The plot shows that californai is sparesly populated, with only one point having a rather heigh `population` cluster. We would further look at how the prices of house a scatter based on location.
sns.relplot(x='latitude', y='longitude', data=df,
hue='price', height=8, palette='jet')
plt.xlabel('latitude',fontsize=16)
plt.ylabel('longitude', fontsize=16)
plt.show()
# #### California's wealthy class seems to reside along the costal region on the state. We would then like to plot the relationship between the location and the `median_income` distribution
sns.relplot(x='latitude',y='longitude', data=df,
hue='price' ,size='median_income', palette='jet', height=10)
plt.show()
# #### Not many houses earn more than $100000 as their `median_income`
df
# #### Does the number of `rooms` have any relationship with the price of the house. let's plot a line graph to check.
sns.relplot('rooms', 'price', data=df,
kind='line', height=6, sort=True)
plt.show()
rooms = df[df['rooms'] <= 20]
df.pivot_table(rooms, 'rooms').iloc[:20,[7]]
# #### From above the more rooms the less the price of the house except for houses with more than 10 rooms, `Note` : Houses with astronomically high number of rooms should be regarded as this may be hotels or caused by mislabelling of data.
# #### houses with `rooms` in the range of 0-20 cost the most.
sns.relplot('bedrooms', 'price', data=df,
height=6, kind='line', sort=True)
plt.show()
rooms = df[df['bedrooms'] <= 20]
df.pivot_table(rooms, 'bedrooms').iloc[:12, [6]]
# #### From above the more bedrooms the less the price of the house except for houses with more than 10 bedrooms, `Note`: Houses wit astronomically high number of bedrooms should be regarded as this may be hotels or caused by mislabelling of data.
sns.relplot('ocean_proximity', 'population', data=df,
kind='line', sort=True,
height=6, palette='jet')
plt.show()
# #### Population distribution of California's per-region, less have island houses, most live around `<1H ocean` region
sns.relplot('ocean_proximity', 'median_income', data=df,
kind='line', sort=True, height=6, palette='jet')
plt.show()
# #### `Island` dwellers have the lowest `median_income` rates, while `<1H ocean` have a very high `median_income` rate.
sns.relplot('ocean_proximity', 'price', data=df,
kind='line', sort=True, height=6, palette='jet')
plt.show()
# #### `Island`houses have the highest `price` rates, while `Inland` house `price` are of very low rate.
sns.relplot('ocean_proximity', 'rooms', data=df,
kind='line', sort=True, height=6, palette='jet')
plt.show()
# #### `Inland` houses have the highest number of `rooms`.
sns.relplot('ocean_proximity', 'housing_median_age', data=df,
kind='line', sort=True, height=6, palette='jet')
plt.show()
# #### `Island`houses have the highest `housing median age`, while `Inland` houses have a `housing_median_age` of `25`.
sns.relplot('ocean_proximity', 'bedrooms', data=df,
kind='line', sort=True, height=6, palette='jet')
plt.show()
# #### `Island`houses have the highest number of `bedrooms`, while `<1H Ocean` and ` Near Bay` have only `1` room.
| 6,710 |
/05_perceptron.ipynb | 2503b70e93925cba896a79963f0765ed8d9bb29b | [] | no_license | Kiana58/NLP_ProgrammingTutorial | https://github.com/Kiana58/NLP_ProgrammingTutorial | 0 | 0 | null | null | null | null | Jupyter Notebook | false | false | .py | 2,944 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Python 2
# language: python
# name: python2
# ---
# +
import math
from collections import defaultdict
def CREATE_FEATURES(x):
phi = defaultdict(int)
words = x.split()
# 1-gram
for word in words:
phi["UNI:"+word] += 1
# 2-gram
for i in xrange(1,len(words)):
phi["BI:"+words[i-1]+" "+words[i]] += 1
return phi
def PREDICT_ONE(w,phi):
score = 0
for name,value in phi.items():
if name in w:
score += value*w[name]
if score>=0:
return 1
else:
return -1
def PREDICT_ALL(model,inp):
w = model
ans = []
for x in inp:
phi = CREATE_FEATURES(x)
y_ = PREDICT_ONE(w,phi)
ans.append(y_)
return ans
def UPDATE_WEIGHTS(w,phi,y):
for name,value in phi.items():
w[name] += value*y
# +
# テスト
# rf = open("../test/03-train-input.txt","r")
# datas = []
# for line in rf.readlines():
# datas.append(line.strip().split("\t"))
# 本番
model_file = open("../data/titles-en-train.labeled","r")
input_file = open("../data/titles-en-test.word","r")
model = model_file.readlines()
inp = input_file.readlines()
datas = []
for line in model:
datas.append(line.strip().split("\t"))
w = defaultdict(int)
for data in datas:
y = float(data[0]); x = data[1]
phi = CREATE_FEATURES(x)
y_ = PREDICT_ONE(w,phi)
if y_ != y:
UPDATE_WEIGHTS(w,phi,y)
with open("../data/my_answer.txt","w") as wf:
for i in PREDICT_ALL(w,inp):
i = str(i)+"\n"
wf.write(i)
# -
# ### UNI-GRAM : Accuracy = 90.967056%
# ### BI-GRAM :Accuracy = 91.321289%
| 1,820 |
/machine-learning-python/machine-learning-chapter-16/assets/subsample.ipynb | bb0d34060d0e1e00111293c48f9f7f84f292f8a8 | [] | no_license | rimjhimroy/katacoda-scenarios-1 | https://github.com/rimjhimroy/katacoda-scenarios-1 | 0 | 1 | null | 2019-11-17T23:19:16 | 2019-11-17T20:05:17 | null | Jupyter Notebook | false | false | .py | 4,545 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: PySpark
# language: ''
# name: pysparkkernel
# ---
# +
# LAB EVALUABLE de SPARK
# -
#import SparkSession
from pyspark.sql import SparkSession
#create spar session object
spark=SparkSession.builder.appName('lab_spark').getOrCreate()
# +
#configuramos python 3
# -
# %%configure -f
{ "conf":{
"spark.pyspark.python": "python3",
"spark.pyspark.virtualenv.enabled": "true",
"spark.pyspark.virtualenv.type":"native",
"spark.pyspark.virtualenv.bin.path":"/usr/bin/virtualenv"
}}
# +
# Basado en la experiencia de haber trabajado con los datos de: datasets/spark/sample_data.csv y
# el notebook: 'Data_processing_using_PySpark.ipynb', realizar un proceso de:
# +
# carga de datos csv en spark desde un bucket S3.
# -
df=spark.read.csv('s3://jscaicedomb/datasets/spark/Casos_positivos_de_COVID-19_en_Colombia.csv',inferSchema=True,header=True)
# +
# borrar y crear algunas columnas
# -
#printSchema
df.printSchema()
#borrar las columnas ID, ciudad, edad, Departamento, codigo, fecha de not y Estado
df_new=df.drop('ID de caso','Ciudad de ubicación','Departamento o Distrito','Codigo DIVIPOLA','Edad','Estado','Fecha de notificación')
df_new.show(10)
#consultamos cuantos hay por ciudad
df.groupBy('Ciudad de ubicación').count().show(5,False)
#crear columnas
# añadimos una columna con la consulta anterior
df.groupBy('Ciudad de ubicación').agg({'Ciudad de ubicación':'count'}).show(5,False)
# +
# realizar filtrados de datos por alguna información que le parezca interesante
# -
# Edad, estado, y fecha de casos en Medellín
df.filter(df['Ciudad de ubicación']=='Medellín').select('Edad','Estado','Fecha de notificación').show()
# Edad, ciudad, y fecha de muerte de fallecidos
df.filter(df['Estado']=='Fallecido').select('Edad','Ciudad de ubicación','Fecha de muerte').show()
#cuantos muertos hay
df.filter(df['Estado']=='Fallecido').count()
# +
# realizar alguna agrupación y consulta de datos categorica, por ejemplo número de casos por región o por sexo/genero.
# -
# Numero de casos por edad
df.groupBy('Edad').count().orderBy('count',ascending=False).show(10,False)
# +
# finalmente grave los resultados en un bucket público en S3
# -
#target directory
write_uri='s3://jscaicedomb/lab_spark'
#save the dataframe as single csv
df.coalesce(1).write.format("csv").option("header","true").save(write_uri)
)
sample_mean = mean([row[0] for row in sample])
sample_means.append(sample_mean)
print('Samples=%d, Estimated Mean: %.3f' % (size, mean(sample_means)))
# Running the above example prints the original mean value we aim to estimate. We can then see
# the estimated mean from the various different numbers of bootstrap samples. We can see that
# with 100 samples we achieve a good estimate of the mean.
| 2,946 |
/listas_practica.ipynb | 39347b80140271f662792196818c7a7ec2fd74db | [] | no_license | Yedi20/ProyectosDataSciences2021 | https://github.com/Yedi20/ProyectosDataSciences2021 | 0 | 0 | null | null | null | null | Jupyter Notebook | false | false | .py | 10,808 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Python (ml)
# language: python
# name: python-ml
# ---
# + [markdown] deletable=true editable=true
# # TensorFlow basics
# + [markdown] deletable=true editable=true
# ## Checking the Installation
# + deletable=true editable=true
>>> import tensorflow as tf
>>> print(tf.__version__)
# + [markdown] deletable=true editable=true
# ## Construction Phase
# + deletable=true editable=true
>>> a = tf.constant(3)
>>> b = tf.constant(5)
>>> s = a + b
# + deletable=true editable=true
a
# + deletable=true editable=true
b
# + deletable=true editable=true
s
# + deletable=true editable=true
tf.get_default_graph()
# + [markdown] deletable=true editable=true
# This code is needed to display TensorFlow graphs in Jupyter:
# + deletable=true editable=true
from IPython.display import display, HTML
def strip_consts(graph_def, max_const_size=32):
"""Strip large constant values from graph_def."""
strip_def = tf.GraphDef()
for n0 in graph_def.node:
n = strip_def.node.add()
n.MergeFrom(n0)
if n.op == 'Const':
tensor = n.attr['value'].tensor
size = len(tensor.tensor_content)
if size > max_const_size:
tensor.tensor_content = b"<stripped %d bytes>"%size
return strip_def
def show_graph(graph_def=None, max_const_size=32):
"""Visualize TensorFlow graph."""
graph_def = graph_def or tf.get_default_graph()
if hasattr(graph_def, 'as_graph_def'):
graph_def = graph_def.as_graph_def()
strip_def = strip_consts(graph_def, max_const_size=max_const_size)
code = """
<script>
function load() {{
document.getElementById("{id}").pbtxt = {data};
}}
</script>
<link rel="import" href="https://tensorboard.appspot.com/tf-graph-basic.build.html" onload=load()>
<div style="height:600px">
<tf-graph-basic id="{id}"></tf-graph-basic>
</div>
""".format(data=repr(str(strip_def)), id='graph'+str(np.random.rand()))
iframe = """
<iframe seamless style="width:1200px;height:620px;border:0" srcdoc="{}"></iframe>
""".format(code.replace('"', '"'))
display(HTML(iframe))
# + deletable=true editable=true
show_graph()
# + deletable=true editable=true
>>> graph = tf.Graph()
>>> with graph.as_default():
... a = tf.constant(3)
... b = tf.constant(5)
... s = a + b
...
# + [markdown] deletable=true editable=true
# ## Execution Phase
# + deletable=true editable=true
>>> with tf.Session(graph=graph) as sess:
... result = s.eval()
...
>>> result
# + deletable=true editable=true
>>> with tf.Session(graph=graph) as sess:
... result = sess.run(s)
...
>>> result
# + deletable=true editable=true
>>> with tf.Session(graph=graph) as sess:
... result = sess.run([a,b,s])
...
>>> result
# + [markdown] deletable=true editable=true
# ## Variables
# + deletable=true editable=true
>>> graph = tf.Graph()
>>> with graph.as_default():
... x = tf.Variable(100)
... c = tf.constant(5)
... increment_op = tf.assign(x, x + c)
...
# + deletable=true editable=true
>>> with tf.Session(graph=graph) as sess:
... x.initializer.run()
... print(x.eval()) # 100
... for iteration in range(10):
... increment_op.eval()
... print(x.eval()) # 150
# + [markdown] deletable=true editable=true
# ## Variables Initializer
# + deletable=true editable=true
>>> graph = tf.Graph()
>>> with graph.as_default():
... x = tf.Variable(100)
... c = tf.constant(5)
... increment_op = tf.assign(x, x + c)
... init = tf.global_variables_initializer()
...
# + deletable=true editable=true
>>> with tf.Session(graph=graph) as sess:
... init.run()
... print(x.eval()) # 100
... for iteration in range(10):
... increment_op.eval()
... print(x.eval()) # 150
# + [markdown] deletable=true editable=true
# ## Variable State
# + deletable=true editable=true
>>> session1 = tf.Session(graph=graph)
>>> session2 = tf.Session(graph=graph)
>>> x.initializer.run(session=session1)
>>> x.initializer.run(session=session2)
# + deletable=true editable=true
>>> increment_op.eval(session=session1)
# + deletable=true editable=true
>>> x.eval(session=session1)
# + deletable=true editable=true
>>> x.eval(session=session2)
# + deletable=true editable=true
>>> session1.close()
>>> session2.close()
# + [markdown] deletable=true editable=true
# ## Collections
# + deletable=true editable=true
>>> graph.get_collection(tf.GraphKeys.GLOBAL_VARIABLES)
# + deletable=true editable=true
tf.GraphKeys.GLOBAL_VARIABLES
# + deletable=true editable=true
>>> graph.add_to_collection("my_collection", c)
>>> graph.get_collection("my_collection")
# + [markdown] deletable=true editable=true
# ## Navigating the Graph
# + deletable=true editable=true
>>> graph = tf.Graph()
>>> with graph.as_default():
... a = tf.constant(3)
... b = tf.constant(5)
... s = a + b
...
>>> graph.get_operations()
# + deletable=true editable=true
>>> graph.get_operation_by_name("add") is s.op
# + deletable=true editable=true
>>> graph.get_tensor_by_name("add:0") is s
# + deletable=true editable=true
>>> list(s.op.inputs)
# + deletable=true editable=true
>>> list(s.op.outputs)
# + [markdown] deletable=true editable=true
# ## Naming Operations
# + deletable=true editable=true
>>> graph = tf.Graph()
>>> with graph.as_default():
... a = tf.constant(3, name='a')
... b = tf.constant(5, name='b')
... s = tf.add(a, b, name='s')
...
# + deletable=true editable=true
>>> graph.get_operations()
# + [markdown] deletable=true editable=true
# # Linear Regression with TensorFlow
# + [markdown] deletable=true editable=true
# ## Loading the training data
# + deletable=true editable=true
import csv
with open('life_satisfaction_vs_gdp_per_capita.csv') as csvfile:
csv_reader = csv.reader(csvfile)
training_data = [record for record in csv_reader]
# + deletable=true editable=true
training_data
# + deletable=true editable=true
import numpy as np
X_train = np.array([[float(record[1])] for record in training_data[1:]]) / 10000 # feature scaling
y_train = np.array([[float(record[2])] for record in training_data[1:]])
# + deletable=true editable=true
X_train
# + deletable=true editable=true
y_train
# + [markdown] deletable=true editable=true
# ## Plot the data
# + deletable=true editable=true
# %matplotlib inline
import matplotlib.pyplot as plt
plt.rcParams['axes.labelsize'] = 14
plt.rcParams['xtick.labelsize'] = 12
plt.rcParams['ytick.labelsize'] = 12
# + deletable=true editable=true
def plot_life_satisfaction(X_train, y_train):
plt.plot(X_train * 10000, y_train, "bo")
plt.axis([0, 60000, 0, 10])
plt.xlabel("GDP per capita ($)")
plt.ylabel("Life Satisfaction")
plt.grid()
plt.figure(figsize=(10,5))
plot_life_satisfaction(X_train, y_train)
plt.show()
# + [markdown] deletable=true editable=true
# ## Building the Linear Regression Model
# + [markdown] deletable=true editable=true
# The predictions are computed using this equation:
#
# $\hat{\mathbf{y}} = \mathbf{X} \cdot \mathbf{w} + b$
#
# + deletable=true editable=true
graph = tf.Graph()
with graph.as_default():
X = tf.constant(X_train, dtype=tf.float32, name="X")
y = tf.constant(y_train, dtype=tf.float32, name="y")
b = tf.Variable(tf.random_uniform([], -1.0, 1.0, seed=42), name="b")
w = tf.Variable(tf.random_uniform([1, 1], -1.0, 1.0, seed=42), name="w")
y_pred = tf.add(tf.matmul(X, w), b, name="y_pred") # X @ w + b
init = tf.global_variables_initializer()
# + deletable=true editable=true
with tf.Session(graph=graph) as sess:
init.run()
print(y_pred.eval())
# + [markdown] deletable=true editable=true
# ## Measuring the Error
# + [markdown] deletable=true editable=true
# The loss function is the Mean Square Error (MSE):
#
# $\text{MSE}(\mathbf{w}, b) = \dfrac{1}{m} \sum\limits_{i=1}^{m}{(\hat{y}^{(i)}-y^{(i)})^2}$
#
# + deletable=true editable=true
with graph.as_default():
error = y_pred - y
square_error = tf.square(error)
mse = tf.reduce_mean(square_error, name="mse")
# + deletable=true editable=true
with tf.Session(graph=graph) as sess:
init.run()
print(mse.eval())
# + [markdown] deletable=true editable=true
# ## Creating the Gradient Descent Operation Manually
# + [markdown] deletable=true editable=true
# The gradient of the MSE with regards to the weight vector $\mathbf{w}$ is:
#
# $\nabla_{\mathbf{w}}\, \text{MSE}(\mathbf{w}, b) =
# \begin{pmatrix}
# \frac{\partial}{\partial w_0} \text{MSE}(\mathbf{w}, b) \\
# \frac{\partial}{\partial w_1} \text{MSE}(\mathbf{w}, b) \\
# \vdots \\
# \frac{\partial}{\partial w_n} \text{MSE}(\mathbf{w}, b)
# \end{pmatrix}
# = \dfrac{2}{m} \mathbf{X}^T \cdot (\hat{\mathbf{y}} - \mathbf{y})
# $
#
# And the partial derivative with regards to the bias $b$ is:
#
# $
# \dfrac{\partial}{\partial b} \text{MSE}(\mathbf{w}, b) = \dfrac{2}{m} \sum\limits_{i=1}^{m}(\hat{y}^{(i)}-y^{(i)})
# $
#
# + deletable=true editable=true
with graph.as_default():
m = tf.cast(tf.shape(X)[0], tf.float32)
gradients_w = 2/m * tf.matmul(tf.transpose(X), error)
gradients_b = 2/m * tf.reduce_sum(error)
# + [markdown] deletable=true editable=true
# Note that `tf.shape(X)` is a tensor that will evaluate to the shape of `X` at runtime, whereas `X.shape` is the build-time shape which may not be fully specified (for example with placeholders, as we will see).
# + [markdown] deletable=true editable=true
# To perform a Gradient Descent step, we need to subtract the gradients (multiplied by the learning rate) from the weight vector and the bias:
#
# $
# \mathbf{w} \gets \mathbf{w} - \eta \nabla_{\mathbf{w}}\, \text{MSE}(\mathbf{w}, b)
# $
#
# $
# \mathbf{b} \gets \mathbf{b} - \eta \dfrac{\partial}{\partial b} \text{MSE}(\mathbf{w}, b)
# $
# + deletable=true editable=true
learning_rate = 0.01
with graph.as_default():
tweak_w_op = tf.assign(w, w - learning_rate * gradients_w)
tweak_b_op = tf.assign(b, b - learning_rate * gradients_b)
training_op = tf.group(tweak_w_op, tweak_b_op)
# + deletable=true editable=true
n_iterations = 2000
with tf.Session(graph=graph) as sess:
init.run()
for iteration in range(n_iterations):
if iteration % 100 == 0:
print("Iteration {:5}, MSE: {:.4f}".format(iteration, mse.eval()))
training_op.run()
w_val, b_val = sess.run([w, b])
# + deletable=true editable=true
def plot_life_satisfaction_with_linear_model(X_train, y_train, w, b):
plot_life_satisfaction(X_train, y_train)
plt.plot([0, 60000], [b, w[0][0] * (60000 / 10000) + b])
plt.figure(figsize=(10, 5))
plot_life_satisfaction_with_linear_model(X_train, y_train, w_val, b_val)
plt.show()
# + [markdown] deletable=true editable=true
# # Using autodiff Instead
# + deletable=true editable=true
graph = tf.Graph()
with graph.as_default():
X = tf.constant(X_train, dtype=tf.float32, name="X")
y = tf.constant(y_train, dtype=tf.float32, name="y")
b = tf.Variable(tf.random_uniform([], -1.0, 1.0, seed=42), name="b")
w = tf.Variable(tf.random_uniform([1, 1], -1.0, 1.0, seed=42), name="w")
y_pred = tf.add(tf.matmul(X, w), b, name="y_pred") # X @ w + b
mse = tf.reduce_mean(tf.square(y_pred - y), name="mse")
gradients_w, gradients_b = tf.gradients(mse, [w, b]) # <= IT'S AUTODIFF MAGIC!
tweak_w_op = tf.assign(w, w - learning_rate * gradients_w)
tweak_b_op = tf.assign(b, b - learning_rate * gradients_b)
training_op = tf.group(tweak_w_op, tweak_b_op)
init = tf.global_variables_initializer()
# + deletable=true editable=true
n_iterations = 2000
with tf.Session(graph=graph) as sess:
init.run()
for iteration in range(n_iterations):
if iteration % 100 == 0:
print("Iteration {:5}, MSE: {:.4f}".format(iteration, mse.eval()))
training_op.run()
w_val, b_val = sess.run([w, b])
# + deletable=true editable=true
plt.figure(figsize=(10, 5))
plot_life_satisfaction_with_linear_model(X_train, y_train, w_val, b_val)
plt.show()
# + [markdown] deletable=true editable=true
# ## Using Optimizers
# + deletable=true editable=true
graph = tf.Graph()
with graph.as_default():
X = tf.constant(X_train, dtype=tf.float32, name="X")
y = tf.constant(y_train, dtype=tf.float32, name="y")
b = tf.Variable(tf.random_uniform([], -1.0, 1.0, seed=42), name="b")
w = tf.Variable(tf.random_uniform([1, 1], -1.0, 1.0, seed=42), name="w")
y_pred = tf.add(tf.matmul(X, w), b, name="y_pred") # X @ w + b
mse = tf.reduce_mean(tf.square(y_pred - y), name="mse")
optimizer = tf.train.GradientDescentOptimizer(learning_rate=learning_rate)
training_op = optimizer.minimize(mse) # <= MOAR AUTODIFF MAGIC!
init = tf.global_variables_initializer()
# + deletable=true editable=true
n_iterations = 2000
with tf.Session(graph=graph) as sess:
init.run()
for iteration in range(n_iterations):
if iteration % 100 == 0:
print("Iteration {:5}, MSE: {:.4f}".format(iteration, mse.eval()))
training_op.run()
w_val, b_val = sess.run([w, b])
# + deletable=true editable=true
plt.figure(figsize=(10, 5))
plot_life_satisfaction_with_linear_model(X_train, y_train, w_val, b_val)
plt.show()
# + [markdown] deletable=true editable=true
# ## Faster Optimizers
# + deletable=true editable=true
momentum = 0.8
graph = tf.Graph()
with graph.as_default():
X = tf.constant(X_train, dtype=tf.float32, name="X")
y = tf.constant(y_train, dtype=tf.float32, name="y")
b = tf.Variable(tf.random_uniform([], -1.0, 1.0, seed=42), name="b")
w = tf.Variable(tf.random_uniform([1, 1], -1.0, 1.0, seed=42), name="w")
y_pred = tf.add(tf.matmul(X, w), b, name="y_pred") # X @ w + b
mse = tf.reduce_mean(tf.square(y_pred - y), name="mse")
optimizer = tf.train.MomentumOptimizer(learning_rate, momentum)
training_op = optimizer.minimize(mse)
init = tf.global_variables_initializer()
# + deletable=true editable=true
n_iterations = 500
with tf.Session(graph=graph) as sess:
init.run()
for iteration in range(n_iterations):
if iteration % 100 == 0:
print("Iteration {:5}, MSE: {:.4f}".format(iteration, mse.eval()))
training_op.run()
w_val, b_val = sess.run([w, b])
# + deletable=true editable=true
plt.figure(figsize=(10, 5))
plot_life_satisfaction_with_linear_model(X_train, y_train, w_val, b_val)
plt.show()
# + [markdown] deletable=true editable=true
# How does the optimizer know which variables to tweak? Answer: the `TRAINABLE_VARIABLES` collection.
# + deletable=true editable=true
coll = graph.get_collection(tf.GraphKeys.TRAINABLE_VARIABLES)
[var.op.name for var in coll]
# + [markdown] deletable=true editable=true
# ## Making Predictions Outside of TensorFlow
# + deletable=true editable=true
cyprus_gdp_per_capita = 22000
cyprus_life_satisfaction = w_val[0][0] * cyprus_gdp_per_capita / 10000 + b_val
cyprus_life_satisfaction
# + [markdown] deletable=true editable=true
# ## Using placeholders
# + deletable=true editable=true
graph = tf.Graph()
with graph.as_default():
X = tf.placeholder(tf.float32, shape=[None, 1], name="X") # <= None allows for any
y = tf.placeholder(tf.float32, shape=[None, 1], name="y") # training batch size
b = tf.Variable(tf.random_uniform([], -1.0, 1.0, seed=42), name="b")
w = tf.Variable(tf.random_uniform([1, 1], -1.0, 1.0, seed=42), name="w")
y_pred = tf.add(tf.matmul(X, w), b, name="y_pred") # X @ w + b
mse = tf.reduce_mean(tf.square(y_pred - y), name="mse")
optimizer = tf.train.MomentumOptimizer(learning_rate, momentum)
training_op = optimizer.minimize(mse)
init = tf.global_variables_initializer()
# + deletable=true editable=true
n_iterations = 500
X_test = np.array([[22000]], dtype=np.float32) / 10000
with tf.Session(graph=graph) as sess:
init.run()
for iteration in range(n_iterations):
feed_dict = {X: X_train, y: y_train}
if iteration % 100 == 0:
print("Iteration {:5}, MSE: {:.4f}".format(
iteration,
mse.eval(feed_dict))) # <= FEED TRAINING DATA
training_op.run(feed_dict) # <= FEED TRAINING DATA
# make the prediction:
y_pred_val = y_pred.eval(feed_dict={X: X_test}) # <= FEED TEST DATA
# + deletable=true editable=true
y_pred_val
# + [markdown] deletable=true editable=true
# ## Saving and Restoring a Model
# + deletable=true editable=true
graph = tf.Graph()
with graph.as_default():
X = tf.placeholder(tf.float32, shape=[None, 1], name="X")
y = tf.placeholder(tf.float32, shape=[None, 1], name="y")
b = tf.Variable(tf.random_uniform([], -1.0, 1.0, seed=42), name="b")
w = tf.Variable(tf.random_uniform([1, 1], -1.0, 1.0, seed=42), name="w")
y_pred = tf.add(tf.matmul(X, w), b, name="y_pred") # X @ w + b
mse = tf.reduce_mean(tf.square(y_pred - y), name="mse")
optimizer = tf.train.MomentumOptimizer(learning_rate, momentum)
training_op = optimizer.minimize(mse)
init = tf.global_variables_initializer()
saver = tf.train.Saver() # <= At the very end of the construction phase
# + deletable=true editable=true
n_iterations = 500
with tf.Session(graph=graph) as sess:
init.run()
for iteration in range(n_iterations):
if iteration % 100 == 0:
print("Iteration {:5}, MSE: {:.4f}".format(
iteration,
mse.eval(feed_dict={X: X_train, y: y_train})))
training_op.run(feed_dict={X: X_train, y: y_train}) # <= FEED THE DICT
saver.save(sess, "./my_life_satisfaction_model")
# + deletable=true editable=true
with tf.Session(graph=graph) as sess:
saver.restore(sess, "./my_life_satisfaction_model")
# make the prediction:
y_pred_val = y_pred.eval(feed_dict={X: X_test})
# + deletable=true editable=true
y_pred_val
# + [markdown] deletable=true editable=true
# ## Restoring a Graph
# + deletable=true editable=true
model_path = "./my_life_satisfaction_model"
graph = tf.Graph()
with tf.Session(graph=graph) as sess:
# restore the graph
saver = tf.train.import_meta_graph(model_path + ".meta")
saver.restore(sess, model_path)
# get references to the tensors we need
X = graph.get_tensor_by_name("X:0")
y_pred = graph.get_tensor_by_name("y_pred:0")
# make the prediction:
y_pred_val = y_pred.eval(feed_dict={X: X_test})
# + deletable=true editable=true
y_pred_val
# + [markdown] deletable=true editable=true
# # Using Readers
# + deletable=true editable=true
filenames = ["life_satisfaction_vs_gdp_per_capita.csv"]
n_epochs = 500
graph = tf.Graph()
with graph.as_default():
reader = tf.TextLineReader(skip_header_lines=1)
filename_queue = tf.train.string_input_producer(filenames, num_epochs=n_epochs)
record_id, record = reader.read(filename_queue)
record_defaults = [[''], [0.0], [0.0]]
country, gdp_per_capita, life_satisfaction = tf.decode_csv(record, record_defaults=record_defaults)
# + deletable=true editable=true
batch_size = 5
with graph.as_default():
X_batch, y_batch = tf.train.batch([gdp_per_capita, life_satisfaction], batch_size=batch_size)
X_batch_reshaped = tf.reshape(X_batch, [-1, 1])
y_batch_reshaped = tf.reshape(y_batch, [-1, 1])
# + deletable=true editable=true
with graph.as_default():
X = tf.placeholder_with_default(X_batch_reshaped, shape=[None, 1], name="X")
y = tf.placeholder_with_default(y_batch_reshaped, shape=[None, 1], name="y")
b = tf.Variable(tf.random_uniform([], -1.0, 1.0, seed=42), name="b")
w = tf.Variable(tf.random_uniform([1, 1], -1.0, 1.0, seed=42), name="w")
y_pred = tf.add(tf.matmul(X / 10000, w), b, name="y_pred") # X @ w + b
mse = tf.reduce_mean(tf.square(y_pred - y), name="mse")
global_step = tf.Variable(0, trainable=False, name='global_step')
optimizer = tf.train.MomentumOptimizer(learning_rate, momentum)
training_op = optimizer.minimize(mse, global_step=global_step)
init = tf.group(tf.global_variables_initializer(), tf.local_variables_initializer())
saver = tf.train.Saver()
# + deletable=true editable=true
with tf.Session(graph=graph) as sess:
init.run()
coord = tf.train.Coordinator()
threads = tf.train.start_queue_runners(coord=coord)
try:
while not coord.should_stop():
_, mse_val, global_step_val = sess.run([training_op, mse, global_step])
if global_step_val % 100 == 0:
print(global_step_val, mse_val)
except tf.errors.OutOfRangeError:
print("End of training")
coord.request_stop()
coord.join(threads)
saver.save(sess, "./my_life_satisfaction_model")
# + [markdown] deletable=true editable=true
# # TensorBoard
# + deletable=true editable=true
from datetime import datetime
def logdir():
root_logdir = "tf_logs"
now = datetime.utcnow().strftime("%Y%m%d%H%M%S")
return "{}/run-{}/".format(root_logdir, now)
summary_writer = tf.summary.FileWriter(logdir(), graph)
# + [markdown] deletable=true editable=true
# Without name scopes, the graph looks like a big mess. So let's add name scopes!
# + deletable=true editable=true
filenames = ["life_satisfaction_vs_gdp_per_capita.csv"]
n_epochs = 500
batch_size = 5
graph = tf.Graph()
with graph.as_default():
with tf.name_scope("reader"):
reader = tf.TextLineReader(skip_header_lines=1)
filename_queue = tf.train.string_input_producer(filenames, num_epochs=n_epochs)
record_id, record = reader.read(filename_queue)
record_defaults = [[''], [0.0], [0.0]]
country, gdp_per_capita, life_satisfaction = tf.decode_csv(record, record_defaults=record_defaults)
X_batch, y_batch = tf.train.batch([gdp_per_capita, life_satisfaction], batch_size=batch_size)
X_batch_reshaped = tf.reshape(X_batch, [-1, 1])
y_batch_reshaped = tf.reshape(y_batch, [-1, 1])
with tf.name_scope("linear_model"):
X = tf.placeholder_with_default(X_batch_reshaped, shape=[None, 1], name="X")
y = tf.placeholder_with_default(y_batch_reshaped, shape=[None, 1], name="y")
b = tf.Variable(tf.random_uniform([], -1.0, 1.0, seed=42), name="b")
w = tf.Variable(tf.random_uniform([1, 1], -1.0, 1.0, seed=42), name="w")
y_pred = tf.add(tf.matmul(X / 10000, w), b, name="y_pred") # X @ w + b
with tf.name_scope("train"):
mse = tf.reduce_mean(tf.square(y_pred - y), name="mse")
global_step = tf.Variable(0, trainable=False, name='global_step')
optimizer = tf.train.MomentumOptimizer(learning_rate, momentum)
training_op = optimizer.minimize(mse, global_step=global_step)
init = tf.group(tf.global_variables_initializer(), tf.local_variables_initializer())
saver = tf.train.Saver()
# + deletable=true editable=true
country.name, gdp_per_capita.name, X_batch.name, y_batch.name
# + deletable=true editable=true
X.name, y.name, b.name, w.name, y_pred.name
# + deletable=true editable=true
mse.name, global_step.name, training_op.name
# + deletable=true editable=true
summary_writer = tf.summary.FileWriter(logdir(), graph)
# + [markdown] deletable=true editable=true
# The graph looks much better in TensorBoard. :)
#
# Okay, now let's add a scalar summary:
# + deletable=true editable=true
filenames = ["life_satisfaction_vs_gdp_per_capita.csv"]
n_epochs = 500
batch_size = 5
graph = tf.Graph()
with graph.as_default():
with tf.name_scope("reader"):
reader = tf.TextLineReader(skip_header_lines=1)
filename_queue = tf.train.string_input_producer(filenames, num_epochs=n_epochs)
record_id, record = reader.read(filename_queue)
record_defaults = [[''], [0.0], [0.0]]
country, gdp_per_capita, life_satisfaction = tf.decode_csv(record, record_defaults=record_defaults)
X_batch, y_batch = tf.train.batch([gdp_per_capita, life_satisfaction], batch_size=batch_size)
X_batch_reshaped = tf.reshape(X_batch, [-1, 1])
y_batch_reshaped = tf.reshape(y_batch, [-1, 1])
with tf.name_scope("linear_model"):
X = tf.placeholder_with_default(X_batch_reshaped, shape=[None, 1], name="X")
y = tf.placeholder_with_default(y_batch_reshaped, shape=[None, 1], name="y")
b = tf.Variable(tf.random_uniform([], -1.0, 1.0, seed=42), name="b")
w = tf.Variable(tf.random_uniform([1, 1], -1.0, 1.0, seed=42), name="w")
y_pred = tf.add(tf.matmul(X / 10000, w), b, name="y_pred") # X @ w + b
with tf.name_scope("train"):
mse = tf.reduce_mean(tf.square(y_pred - y), name="mse")
mse_summary = tf.summary.scalar('MSE', mse)
global_step = tf.Variable(0, trainable=False, name='global_step')
optimizer = tf.train.MomentumOptimizer(learning_rate, momentum)
training_op = optimizer.minimize(mse, global_step=global_step)
init = tf.group(tf.global_variables_initializer(), tf.local_variables_initializer())
saver = tf.train.Saver()
# + deletable=true editable=true
summary_writer = tf.summary.FileWriter(logdir(), graph)
# + [markdown] deletable=true editable=true
# And during training, let's write these summaries to the logs:
# + deletable=true editable=true
with tf.Session(graph=graph) as sess:
init.run()
coord = tf.train.Coordinator()
threads = tf.train.start_queue_runners(coord=coord)
try:
while not coord.should_stop():
_, mse_summary_val, global_step_val = sess.run([training_op, mse_summary, global_step])
if global_step_val % 10 == 0:
summary_writer.add_summary(mse_summary_val, global_step_val)
except tf.errors.OutOfRangeError:
print("End of training")
coord.request_stop()
coord.join(threads)
saver.save(sess, "./my_life_satisfaction_model")
# + [markdown] deletable=true editable=true
# # Artificial Neural Networks
# + [markdown] deletable=true editable=true
# Load MNIST dataset:
# + deletable=true editable=true
from tensorflow.examples.tutorials.mnist import input_data
mnist = input_data.read_data_sets("/tmp/data/")
# + deletable=true editable=true
batch_size = 3
X_batch, y_batch = mnist.train.next_batch(batch_size)
X_batch.shape
# + deletable=true editable=true
for image_data in X_batch:
plt.imshow(image_data.reshape([28, 28]), cmap="binary", interpolation="nearest")
plt.show()
# + deletable=true editable=true
y_batch
# + [markdown] deletable=true editable=true
# Build the model:
# + deletable=true editable=true
n_inputs = 28*28
n_hidden1 = 100
n_outputs = 10
graph = tf.Graph()
with graph.as_default():
with tf.name_scope("inputs"):
X = tf.placeholder(tf.float32, shape=[None, n_inputs], name="X")
y = tf.placeholder(tf.int32, shape=[None], name="y")
with tf.name_scope("hidden1"):
b1 = tf.Variable(tf.random_uniform([n_hidden1], -1.0, 1.0, seed=42), name="b1")
W1 = tf.Variable(tf.random_uniform([n_inputs, n_hidden1], -1.0, 1.0, seed=42), name="W1")
hidden1 = tf.nn.relu(tf.matmul(X, W1) + b1)
with tf.name_scope("output"):
b2 = tf.Variable(tf.random_uniform([n_outputs], -1.0, 1.0, seed=42), name="b2")
W2 = tf.Variable(tf.random_uniform([n_hidden1, n_outputs], -1.0, 1.0, seed=42), name="W2")
logits = tf.matmul(hidden1, W2) + b2
Y_proba = tf.nn.softmax(logits, name="Y_proba")
with tf.name_scope("train"):
xentropy = tf.nn.sparse_softmax_cross_entropy_with_logits(logits=logits, labels=y)
loss = tf.reduce_mean(xentropy)
optimizer = tf.train.AdamOptimizer()
training_op = optimizer.minimize(loss)
with tf.name_scope("eval"):
correct = tf.nn.in_top_k(logits, y, 1)
accuracy = tf.reduce_mean(tf.cast(correct, tf.float32))
with tf.name_scope("init_and_save"):
init = tf.global_variables_initializer()
saver = tf.train.Saver()
# + [markdown] deletable=true editable=true
# Train the model:
# + deletable=true editable=true
n_epochs = 20
batch_size = 50
with tf.Session(graph=graph) as sess:
init.run()
for epoch in range(n_epochs):
for iteration in range(mnist.train.num_examples // batch_size):
X_batch, y_batch = mnist.train.next_batch(batch_size)
sess.run(training_op, feed_dict={X: X_batch, y: y_batch})
acc_train = accuracy.eval(feed_dict={X: X_batch, y: y_batch})
acc_test = accuracy.eval(feed_dict={X: mnist.test.images, y: mnist.test.labels})
print(epoch, "Train accuracy:", acc_train, "Test accuracy:", acc_test)
save_path = saver.save(sess, "./my_mnist_model")
# + [markdown] deletable=true editable=true
# Load the model and make predictions:
# + deletable=true editable=true
graph = tf.Graph()
with tf.Session(graph=graph) as sess:
saver = tf.train.import_meta_graph("./my_mnist_model.meta")
saver.restore(sess, "./my_mnist_model")
X = graph.get_tensor_by_name("inputs/X:0")
Y_proba = graph.get_tensor_by_name("output/Y_proba:0")
Y_proba_val = Y_proba.eval(feed_dict={X: mnist.test.images})
# + deletable=true editable=true
for example_index in range(200):
y_proba = Y_proba_val[example_index]
y_pred = np.argmax(y_proba)
y_label = mnist.test.labels[example_index]
print("Example {}: prediction={}, label={}, correct={}".format(example_index, y_pred, y_label, y_pred==y_label))
if y_pred != y_label:
print(" Probabilities:", " ".join(["{}:{:.1f}%".format(n, 100*p)
for n, p in enumerate(y_proba) if p > 0.01]))
plt.imshow(mnist.test.images[example_index].reshape([28, 28]), cmap="binary", interpolation="nearest")
plt.show()
# + [markdown] deletable=true editable=true
# # Organizing Your Code
# + deletable=true editable=true
def neural_net_layer(inputs, n_neurons, activation=None, seed=None):
n_inputs = int(inputs.get_shape()[1])
b = tf.Variable(tf.random_uniform([n_neurons], -1.0, 1.0, seed=seed), name="b")
W = tf.Variable(tf.random_uniform([n_inputs, n_neurons], -1.0, 1.0, seed=seed), name="W")
logits = tf.matmul(inputs, W) + b
if activation:
return activation(logits)
else:
return logits
# + [markdown] deletable=true editable=true
# Let's simplify our code by using `neural_net_layer()`:
# + deletable=true editable=true
n_inputs = 28*28
n_hidden1 = 100
n_outputs = 10
graph = tf.Graph()
with graph.as_default():
with tf.name_scope("inputs"):
X = tf.placeholder(tf.float32, shape=[None, n_inputs], name="X")
y = tf.placeholder(tf.int32, shape=[None], name="y")
#########################################################################
# This section is simplified (the rest is unchanged)
#
with tf.name_scope("hidden1"):
hidden1 = neural_net_layer(X, n_hidden1, activation=tf.nn.relu) # <= CHANGED
with tf.name_scope("output"):
logits = neural_net_layer(hidden1, n_outputs) # <= CHANGED
Y_proba = tf.nn.softmax(logits, name="Y_proba")
#
#
#########################################################################
with tf.name_scope("train"):
xentropy = tf.nn.sparse_softmax_cross_entropy_with_logits(logits=logits, labels=y)
loss = tf.reduce_mean(xentropy)
optimizer = tf.train.AdamOptimizer()
training_op = optimizer.minimize(loss)
with tf.name_scope("eval"):
correct = tf.nn.in_top_k(logits, y, 1)
accuracy = tf.reduce_mean(tf.cast(correct, tf.float32))
with tf.name_scope("init_and_save"):
init = tf.global_variables_initializer()
saver = tf.train.Saver()
# + deletable=true editable=true
[var.op.name for var in graph.get_collection(tf.GraphKeys.TRAINABLE_VARIABLES)]
# + [markdown] deletable=true editable=true
# Let's check that training still works:
# + deletable=true editable=true
n_epochs = 20
batch_size = 50
with tf.Session(graph=graph) as sess:
init.run()
for epoch in range(n_epochs):
for iteration in range(mnist.train.num_examples // batch_size):
X_batch, y_batch = mnist.train.next_batch(batch_size)
sess.run(training_op, feed_dict={X: X_batch, y: y_batch})
acc_train = accuracy.eval(feed_dict={X: X_batch, y: y_batch})
acc_test = accuracy.eval(feed_dict={X: mnist.test.images, y: mnist.test.labels})
print(epoch, "Train accuracy:", acc_train, "Test accuracy:", acc_test)
save_path = saver.save(sess, "./my_mnist_model")
# + [markdown] deletable=true editable=true
# Now let's use `tf.layers.dense()` instead:
# + deletable=true editable=true
n_inputs = 28*28
n_hidden1 = 100
n_outputs = 10
graph = tf.Graph()
with graph.as_default():
with tf.name_scope("inputs"):
X = tf.placeholder(tf.float32, shape=[None, n_inputs], name="X")
y = tf.placeholder(tf.int32, shape=[None], name="y")
with tf.name_scope("hidden1"):
hidden1 = tf.layers.dense(X, n_hidden1, activation=tf.nn.relu, name="hidden1") # <= CHANGED
with tf.name_scope("output"):
logits = tf.layers.dense(hidden1, n_outputs, name="output") # <= CHANGED
Y_proba = tf.nn.softmax(logits)
with tf.name_scope("train"):
xentropy = tf.nn.sparse_softmax_cross_entropy_with_logits(logits=logits, labels=y)
loss = tf.reduce_mean(xentropy)
optimizer = tf.train.AdamOptimizer()
training_op = optimizer.minimize(loss)
with tf.name_scope("eval"):
correct = tf.nn.in_top_k(logits, y, 1)
accuracy = tf.reduce_mean(tf.cast(correct, tf.float32))
with tf.name_scope("init_and_save"):
init = tf.global_variables_initializer()
saver = tf.train.Saver()
# + deletable=true editable=true
[var.op.name for var in graph.get_collection(tf.GraphKeys.TRAINABLE_VARIABLES)]
# + [markdown] deletable=true editable=true
# Let's check that training still works:
# + deletable=true editable=true
n_epochs = 20
batch_size = 50
with tf.Session(graph=graph) as sess:
init.run()
for epoch in range(n_epochs):
for iteration in range(mnist.train.num_examples // batch_size):
X_batch, y_batch = mnist.train.next_batch(batch_size)
sess.run(training_op, feed_dict={X: X_batch, y: y_batch})
acc_train = accuracy.eval(feed_dict={X: X_batch, y: y_batch})
acc_test = accuracy.eval(feed_dict={X: mnist.test.images, y: mnist.test.labels})
print(epoch, "Train accuracy:", acc_train, "Test accuracy:", acc_test)
save_path = saver.save(sess, "./my_mnist_model")
# + [markdown] deletable=true editable=true
# Now suppose you want two more hidden layers with shared weights & biases. Let's use variable scopes for this:
# + deletable=true editable=true
n_inputs = 28*28
n_hidden = 100
n_outputs = 10
graph = tf.Graph()
with graph.as_default():
with tf.name_scope("inputs"):
X = tf.placeholder(tf.float32, shape=[None, n_inputs], name="X")
y = tf.placeholder(tf.int32, shape=[None], name="y")
hidden1 = tf.layers.dense(X, n_hidden, activation=tf.nn.relu, name="hidden1") # <= CHANGED
hidden2 = tf.layers.dense(hidden1, n_hidden, activation=tf.nn.relu, name="hidden23") # <= CHANGED
hidden3 = tf.layers.dense(hidden2, n_hidden, activation=tf.nn.relu, name="hidden23", reuse=True) # <= CHANGED
with tf.name_scope("output"):
logits = tf.layers.dense(hidden3, n_outputs, name="output")
Y_proba = tf.nn.softmax(logits, name="Y_proba")
with tf.name_scope("train"):
xentropy = tf.nn.sparse_softmax_cross_entropy_with_logits(logits=logits, labels=y)
loss = tf.reduce_mean(xentropy)
optimizer = tf.train.AdamOptimizer()
training_op = optimizer.minimize(loss)
with tf.name_scope("eval"):
correct = tf.nn.in_top_k(logits, y, 1)
accuracy = tf.reduce_mean(tf.cast(correct, tf.float32))
with tf.name_scope("init_and_save"):
init = tf.global_variables_initializer()
saver = tf.train.Saver()
# + deletable=true editable=true
[var.op.name for var in graph.get_collection(tf.GraphKeys.TRAINABLE_VARIABLES)]
# + [markdown] deletable=true editable=true
# Check that training works well:
# + deletable=true editable=true
n_epochs = 20
batch_size = 50
with tf.Session(graph=graph) as sess:
init.run()
for epoch in range(n_epochs):
for iteration in range(mnist.train.num_examples // batch_size):
X_batch, y_batch = mnist.train.next_batch(batch_size)
sess.run(training_op, feed_dict={X: X_batch, y: y_batch})
acc_train = accuracy.eval(feed_dict={X: X_batch, y: y_batch})
acc_test = accuracy.eval(feed_dict={X: mnist.test.images, y: mnist.test.labels})
print(epoch, "Train accuracy:", acc_train, "Test accuracy:", acc_test)
save_path = saver.save(sess, "./my_mnist_model")
# + [markdown] deletable=true editable=true
# How would we implement variable sharing in `neural_net_layer()`?
# + deletable=true editable=true
def neural_net_layer(inputs, n_neurons, activation=None, name=None, reuse=None, seed=None):
with tf.variable_scope(name, default_name="layer", reuse=reuse):
n_inputs = int(inputs.get_shape()[1])
rnd_init = lambda shape, dtype, partition_info: tf.random_uniform(shape, -1.0, 1.0, dtype=dtype, seed=seed)
b = tf.get_variable("biases", shape=[n_neurons], initializer=rnd_init)
W = tf.get_variable("weights", shape=[n_inputs, n_neurons], initializer=rnd_init)
logits = tf.matmul(inputs, W) + b
if activation:
return activation(logits)
else:
return logits
# + deletable=true editable=true
graph = tf.Graph()
with graph.as_default():
with tf.variable_scope("foo"):
a = tf.constant(1., name="a")
with tf.name_scope("bar"):
b = tf.constant(2., name="b")
with tf.name_scope("baz"):
c = tf.get_variable("c", shape=[], initializer=tf.constant_initializer(2))
s = tf.add_n([a,b,c], name="s")
# + deletable=true editable=true
a.name
# + deletable=true editable=true
b.name
# + deletable=true editable=true
c.name
# + deletable=true editable=true
s.name
# + [markdown] deletable=true editable=true
# # Techniques for Training Deep Nets
# + [markdown] deletable=true editable=true
# Using He initialization and the ELU activation function (with the help of a `partial()`):
# + deletable=true editable=true
from functools import partial
n_inputs = 28*28
n_hidden1 = 100
n_hidden2 = 100
n_outputs = 10
graph = tf.Graph()
with graph.as_default():
with tf.name_scope("inputs"):
X = tf.placeholder(tf.float32, shape=[None, n_inputs], name="X")
y = tf.placeholder(tf.int32, shape=[None], name="y")
he_init = tf.contrib.layers.variance_scaling_initializer()
dense_layer = partial(tf.layers.dense,
kernel_initializer=he_init,
activation=tf.nn.elu)
hidden1 = dense_layer(X, n_hidden1, name="hidden1")
hidden2 = dense_layer(hidden1, n_hidden2, name="hidden2")
logits = dense_layer(hidden2, n_outputs, activation=None, name="output")
Y_proba = tf.nn.softmax(logits)
with tf.name_scope("train"):
xentropy = tf.nn.sparse_softmax_cross_entropy_with_logits(logits=logits, labels=y)
loss = tf.reduce_mean(xentropy)
optimizer = tf.train.AdamOptimizer()
training_op = optimizer.minimize(loss)
with tf.name_scope("eval"):
correct = tf.nn.in_top_k(logits, y, 1)
accuracy = tf.reduce_mean(tf.cast(correct, tf.float32))
with tf.name_scope("init_and_save"):
init = tf.global_variables_initializer()
saver = tf.train.Saver()
# + deletable=true editable=true
n_epochs = 20
batch_size = 50
with tf.Session(graph=graph) as sess:
init.run()
for epoch in range(n_epochs):
for iteration in range(mnist.train.num_examples // batch_size):
X_batch, y_batch = mnist.train.next_batch(batch_size)
sess.run(training_op, feed_dict={X: X_batch, y: y_batch})
acc_train = accuracy.eval(feed_dict={X: X_batch, y: y_batch})
acc_test = accuracy.eval(feed_dict={X: mnist.test.images, y: mnist.test.labels})
print(epoch, "Train accuracy:", acc_train, "Test accuracy:", acc_test)
save_path = saver.save(sess, "./my_mnist_model")
# + [markdown] deletable=true editable=true
# Let's try with 50% dropout now:
# + deletable=true editable=true
n_inputs = 28*28
n_hidden1 = 100
n_hidden2 = 100
n_outputs = 10
dropout_rate = 0.5 # <= CHANGED
graph = tf.Graph()
with graph.as_default():
with tf.name_scope("inputs"):
X = tf.placeholder(tf.float32, shape=[None, n_inputs], name="X")
y = tf.placeholder(tf.int32, shape=[None], name="y")
training = tf.placeholder_with_default(False, shape=[], name='training') # <= CHANGED
X_drop = tf.layers.dropout(X, dropout_rate, training=training) # <= CHANGED
he_init = tf.contrib.layers.variance_scaling_initializer()
dense_layer = partial(tf.layers.dense,
kernel_initializer=he_init,
activation=tf.nn.elu)
hidden1 = dense_layer(X_drop, n_hidden1, name="hidden1") # <= CHANGED
hidden2 = dense_layer(hidden1, n_hidden2, name="hidden2")
logits = dense_layer(hidden2, n_outputs, activation=None, name="output")
Y_proba = tf.nn.softmax(logits)
with tf.name_scope("train"):
xentropy = tf.nn.sparse_softmax_cross_entropy_with_logits(logits=logits, labels=y)
loss = tf.reduce_mean(xentropy)
optimizer = tf.train.AdamOptimizer()
training_op = optimizer.minimize(loss)
with tf.name_scope("eval"):
correct = tf.nn.in_top_k(logits, y, 1)
accuracy = tf.reduce_mean(tf.cast(correct, tf.float32))
with tf.name_scope("init_and_save"):
init = tf.global_variables_initializer()
saver = tf.train.Saver()
# + [markdown] deletable=true editable=true
# How does this model perform?
# + deletable=true editable=true
n_epochs = 20
batch_size = 50
with tf.Session(graph=graph) as sess:
init.run()
for epoch in range(n_epochs):
for iteration in range(mnist.train.num_examples // batch_size):
X_batch, y_batch = mnist.train.next_batch(batch_size)
sess.run(training_op, feed_dict={X: X_batch, y: y_batch, training: True}) # <= CHANGED
acc_train = accuracy.eval(feed_dict={X: X_batch, y: y_batch})
acc_test = accuracy.eval(feed_dict={X: mnist.test.images, y: mnist.test.labels})
print(epoch, "Train accuracy:", acc_train, "Test accuracy:", acc_test)
save_path = saver.save(sess, "./my_mnist_model")
# + [markdown] deletable=true editable=true
# Let's use early stopping:
# + deletable=true editable=true
n_epochs = 1000
batch_size = 50
best_acc_val = 0
check_interval = 100
checks_since_last_progress = 0
max_checks_without_progress = 100
with tf.Session(graph=graph) as sess:
init.run()
for epoch in range(n_epochs):
for iteration in range(mnist.train.num_examples // batch_size):
X_batch, y_batch = mnist.train.next_batch(batch_size)
sess.run(training_op, feed_dict={X: X_batch, y: y_batch, training: True})
if iteration % check_interval == 0:
acc_val = accuracy.eval(feed_dict={X: mnist.test.images[:2000], y: mnist.test.labels[:2000]})
if acc_val > best_acc_val:
best_acc_val = acc_val
checks_since_last_progress = 0
saver.save(sess, "./my_best_model_so_far")
else:
checks_since_last_progress += 1
acc_train = accuracy.eval(feed_dict={X: X_batch, y: y_batch})
acc_test = accuracy.eval(feed_dict={X: mnist.test.images[2000:], y: mnist.test.labels[2000:]})
print(epoch, "Train accuracy:", acc_train, "Test accuracy:", acc_test, "Best validation accuracy:", best_acc_val)
if checks_since_last_progress > max_checks_without_progress:
print("Early stopping!")
saver.restore(sess, "./my_best_model_so_far")
break
acc_test = accuracy.eval(feed_dict={X: mnist.test.images[2000:], y: mnist.test.labels[2000:]})
print("Final accuracy on test set:", acc_test)
save_path = saver.save(sess, "./my_mnist_model")
# + [markdown] deletable=true editable=true
# Saving the model to disk so often slows down training. Let's save to RAM instead:
# + deletable=true editable=true
def get_model_params():
gvars = tf.get_collection(tf.GraphKeys.GLOBAL_VARIABLES)
return {gvar.op.name: value for gvar, value in zip(gvars, tf.get_default_session().run(gvars))}
def restore_model_params(model_params):
gvar_names = list(model_params.keys())
assign_ops = {gvar_name: tf.get_default_graph().get_operation_by_name(gvar_name + "/Assign")
for gvar_name in gvar_names}
init_values = {gvar_name: assign_op.inputs[1] for gvar_name, assign_op in assign_ops.items()}
feed_dict = {init_values[gvar_name]: model_params[gvar_name] for gvar_name in gvar_names}
tf.get_default_session().run(assign_ops, feed_dict=feed_dict)
# + deletable=true editable=true
n_epochs = 1000
batch_size = 50
best_acc_val = 0
check_interval = 100
checks_since_last_progress = 0
max_checks_without_progress = 100
best_model_params = None
with tf.Session(graph=graph) as sess:
init.run()
for epoch in range(n_epochs):
for iteration in range(mnist.train.num_examples // batch_size):
X_batch, y_batch = mnist.train.next_batch(batch_size)
sess.run(training_op, feed_dict={X: X_batch, y: y_batch, training: True})
if iteration % check_interval == 0:
acc_val = accuracy.eval(feed_dict={X: mnist.test.images[:2000], y: mnist.test.labels[:2000]})
if acc_val > best_acc_val:
best_acc_val = acc_val
checks_since_last_progress = 0
best_model_params = get_model_params()
else:
checks_since_last_progress += 1
acc_train = accuracy.eval(feed_dict={X: X_batch, y: y_batch})
acc_test = accuracy.eval(feed_dict={X: mnist.test.images[2000:], y: mnist.test.labels[2000:]})
print(epoch, "Train accuracy:", acc_train, "Test accuracy:", acc_test, "Best validation accuracy:", best_acc_val)
if checks_since_last_progress > max_checks_without_progress:
print("Early stopping!")
break
if best_model_params:
restore_model_params(best_model_params)
acc_test = accuracy.eval(feed_dict={X: mnist.test.images[2000:], y: mnist.test.labels[2000:]})
print("Final accuracy on test set:", acc_test)
save_path = saver.save(sess, "./my_mnist_model")
# + [markdown] deletable=true editable=true
# # Convolutional Neural Networks
# + [markdown] deletable=true editable=true
# Load demo image:
# + deletable=true editable=true
from scipy.misc import imread
china = imread("./images/china.png")
# + deletable=true editable=true
china.shape
# + deletable=true editable=true
def plot_image(image):
cmap = "gray" if len(image.shape) == 2 else None
plt.imshow(image, cmap=cmap, interpolation="nearest")
plt.axis("off")
# + deletable=true editable=true
plt.figure(figsize=(10,7))
plot_image(china)
# + [markdown] deletable=true editable=true
# Crop it and convert it to grayscale:
# + deletable=true editable=true
image = china[150:220, 130:250].mean(axis=2).astype(np.float32)
image.shape
# + deletable=true editable=true
height, width = image.shape
channels = 1 # grayscale
# + deletable=true editable=true
plt.figure(figsize=(10,6))
plot_image(image)
# + deletable=true editable=true
basic_filters = np.zeros(shape=(7, 7, 1, 2), dtype=np.float32) # height, width, in channels, out channels
basic_filters[:, 3, 0, 0] = 1
basic_filters[3, :, 0, 1] = 1
plot_image(basic_filters[:, :, 0, 0])
plt.show()
plot_image(basic_filters[:, :, 0, 1])
plt.show()
# + deletable=true editable=true
graph = tf.Graph()
with graph.as_default():
X = tf.placeholder(tf.float32, shape=(None, height, width, channels))
filters = tf.constant(basic_filters)
convolution = tf.nn.conv2d(X, filters, strides=[1,1,1,1], padding="SAME")
# + deletable=true editable=true
with tf.Session(graph=graph) as sess:
X_batch = image.reshape(1, height, width, 1)
output = convolution.eval(feed_dict={X: X_batch})
# + deletable=true editable=true
plt.figure(figsize=(10,6))
plot_image(output[0, :, :, 0])
# + deletable=true editable=true
plt.figure(figsize=(10,6))
plot_image(output[0, :, :, 1])
# + [markdown] deletable=true editable=true
# Now let's add a max pooling layer:
# + deletable=true editable=true
graph = tf.Graph()
with graph.as_default():
X = tf.placeholder(tf.float32, shape=(None, height, width, channels))
filters = tf.constant(basic_filters)
convolution = tf.nn.conv2d(X, filters, strides=[1,1,1,1], padding="SAME")
max_pool = tf.nn.max_pool(convolution, ksize=[1, 2, 2, 1], strides=[1, 2, 2, 1], padding="VALID")
# + deletable=true editable=true
with tf.Session(graph=graph) as sess:
X_batch = image.reshape(1, height, width, 1)
output = max_pool.eval(feed_dict={X: X_batch})
# + deletable=true editable=true
plt.figure(figsize=(5,3))
plot_image(output[0, :, :, 0])
# + deletable=true editable=true
plt.figure(figsize=(5,3))
plot_image(output[0, :, :, 1])
# + [markdown] deletable=true editable=true
# Let's tackle MNIST using the whole deal:
#
# * Two convolutional layers (using 3x3 filters, SAME padding and stride 1) followed by a max pooling layer, with 32 and 64 feature maps.
# * One Fully Connected (FC) layer with 128 neurons
# * A Softmax output layer to classify images in the 10 classes
# * Applying Dropout after the max pooling layer and after the FC layer.
# * As usual, we optimize the cross-entropy, using an Adam optimizer.
# + deletable=true editable=true
height = 28
width = 28
channels = 1
conv1_fmaps = 32
conv1_ksize = 3
conv1_stride = 1
conv1_pad = "SAME"
conv2_fmaps = 64
conv2_ksize = 3
conv2_stride = 1
conv2_pad = "SAME"
conv2_dropout_rate = 0.25
pool3_fmaps = conv2_fmaps
n_fc1 = 128
fc1_dropout_rate = 0.5
n_outputs = 10
graph = tf.Graph()
with graph.as_default():
with tf.name_scope("inputs"):
X = tf.placeholder(tf.float32, shape=[None, n_inputs], name="X")
X_reshaped = tf.reshape(X, shape=[-1, height, width, channels])
y = tf.placeholder(tf.int32, shape=[None], name="y")
training = tf.placeholder_with_default(False, shape=[], name='training')
conv1 = tf.layers.conv2d(X_reshaped, conv1_fmaps, kernel_size=conv1_ksize, strides=conv1_stride, padding=conv1_pad, activation=tf.nn.relu, name="conv1")
conv2 = tf.layers.conv2d(conv1, conv2_fmaps, kernel_size=conv2_ksize, strides=conv2_stride, padding=conv2_pad, activation=tf.nn.relu, name="conv2")
with tf.name_scope("pool3"):
pool3 = tf.nn.max_pool(conv2, ksize=[1, 2, 2, 1], strides=[1, 2, 2, 1], padding="VALID")
pool3_flat = tf.reshape(pool3, shape=[-1, pool3_fmaps * 14 * 14])
pool3_flat_drop = tf.layers.dropout(pool3_flat, conv2_dropout_rate, training=training)
with tf.name_scope("fc1"):
fc1 = tf.layers.dense(pool3_flat_drop, n_fc1, activation=tf.nn.relu, name="fc1")
fc1_drop = tf.layers.dropout(fc1, fc1_dropout_rate, training=training)
with tf.name_scope("output"):
logits = tf.layers.dense(fc1, n_outputs, name="output")
Y_proba = tf.nn.softmax(logits, name="Y_proba")
with tf.name_scope("train"):
xentropy = tf.nn.sparse_softmax_cross_entropy_with_logits(logits=logits, labels=y)
loss = tf.reduce_mean(xentropy)
optimizer = tf.train.AdamOptimizer()
training_op = optimizer.minimize(loss)
with tf.name_scope("eval"):
correct = tf.nn.in_top_k(logits, y, 1)
accuracy = tf.reduce_mean(tf.cast(correct, tf.float32))
with tf.name_scope("init_and_save"):
init = tf.global_variables_initializer()
saver = tf.train.Saver()
# + [markdown] deletable=true editable=true
# Now let training begin, using early stopping. This is quite slow on a CPU, but much faster on a GPU. We achieve >99% accuracy on the test set.
# + deletable=true editable=true
n_epochs = 1000
batch_size = 50
best_acc_val = 0
check_interval = 100
checks_since_last_progress = 0
max_checks_without_progress = 100
best_model_params = None
with tf.Session(graph=graph) as sess:
init.run()
for epoch in range(n_epochs):
for iteration in range(mnist.train.num_examples // batch_size):
X_batch, y_batch = mnist.train.next_batch(batch_size)
sess.run(training_op, feed_dict={X: X_batch, y: y_batch, training: True})
if iteration % check_interval == 0:
acc_val = accuracy.eval(feed_dict={X: mnist.test.images[:2000], y: mnist.test.labels[:2000]})
if acc_val > best_acc_val:
best_acc_val = acc_val
checks_since_last_progress = 0
best_model_params = get_model_params()
else:
checks_since_last_progress += 1
acc_train = accuracy.eval(feed_dict={X: X_batch, y: y_batch})
acc_test = accuracy.eval(feed_dict={X: mnist.test.images[2000:], y: mnist.test.labels[2000:]})
print(epoch, "Train accuracy:", acc_train, "Test accuracy:", acc_test, "Best validation accuracy:", best_acc_val)
if checks_since_last_progress > max_checks_without_progress:
print("Early stopping!")
break
if best_model_params:
restore_model_params(best_model_params)
acc_test = accuracy.eval(feed_dict={X: mnist.test.images[2000:], y: mnist.test.labels[2000:]})
print("Final accuracy on test set:", acc_test)
save_path = saver.save(sess, "./my_mnist_model")
# + deletable=true editable=true
with tf.Session(graph=graph) as sess:
init.run()
saver.restore(sess, "./my_mnist_model")
Y_proba_val = Y_proba.eval(feed_dict={X: mnist.test.images[2000:2400]})
# + deletable=true editable=true
for image, y_label, y_proba in zip(mnist.test.images[2000:2400], mnist.test.labels[2000:2400], Y_proba_val):
y_pred = np.argmax(y_proba)
if y_pred != y_label or y_proba[y_label] < 0.8:
print("Label: {}, Prediction: {}, Probabilities: {}".format(
y_label, y_pred,
" ".join(["{}={:.1f}%".format(n, 100*p)
for n, p in enumerate(y_proba) if p > 0.01])))
plt.imshow(image.reshape(28, 28), cmap="binary")
plt.axis("off")
plt.show()
# + deletable=true editable=true
| 55,272 |
/Capstone project.ipynb | 6dcfc9bed559319cd40d3d5fb5be8bf65ae7b192 | [] | no_license | SamrachTen/Capstone-Project | https://github.com/SamrachTen/Capstone-Project | 0 | 0 | null | null | null | null | Jupyter Notebook | false | false | .py | 1,738 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Python 3.6
# language: python
# name: python3
# ---
# # Capstone Project
# ## This notebook will be mainly used for the capstone project
import pandas as pd
import numpy as np
print("Hello Capstone Project Course!")
xis="month"
# z-axis(color)="passengers"
######
# -
# Perform imports here:
import plotly.offline as pyo
import plotly.graph_objs as go
import pandas as pd
# Create a DataFrame from "flights" data
df = pd.read_csv('../__data/flights.csv')
df.head()
# Define a data variable
data = [go.Heatmap(x=df['year'], y=df['month'], z=df['passengers'])]
# Define the layout
layout = go.Layout(title='Flights', xaxis=dict(title='Year'), yaxis=dict(title='Month'))
# +
# Create a fig from data and layout, and plot the fig
fig = go.Figure(data=data, layout=layout)
pyo.plot(fig, filename='Heatmaps Exercise.html')
# -
| 1,052 |
/homework/Day_065_HW.ipynb | 657329fd14414cb03845890a02b9d631410fc8bf | [] | no_license | jim60304ko/2nd-ML100Days | https://github.com/jim60304ko/2nd-ML100Days | 0 | 0 | null | null | null | null | Jupyter Notebook | false | false | .py | 8,244 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Summary
#
# The long term rainfall data (between 1994 and 2018) at the 4 major reservoirs were plotted and examined for any trends. Plots of the monthly, annual, maximum, and minimum values showed no significant deviations from "normal" rainfall patterns. However, in the plot of dry season and wet season rainfall at Navet reservoir, rainfall was shown to be decreasing. It is recommended that this study be conducted with a longer dataset if possible.
# # Aim
# There's been a lot of speculation that rainfall has been decreasing over the years, causing water woes throughout Trinidad and Tobago. Here I will examine the historical monthly rainfall values at the 4 major reservoirs (Caroni-Arena, Hollis, Navet, and Hillsborough in Tobago) to look for any evidence that this is true. I have months from 1994 to 2018. That's not enough but let's go anyway.
import pandas as pd
# %matplotlib inline
# # Data Prep
df = pd.read_csv("ReservoirRainfall.csv")
df.Date = pd.to_datetime(df.Date) #Changing the date column to date objects and setting as the index
df = df.sort_values('Date')
df.set_index('Date', inplace=True)
df.head()
df.describe() #won't bother checking for nulls because I know there aren't any. Poor practice I know yes
# From the description of the reservoirs, everything looks as expected. Hollis clearly gets more rainfall than everybody else. I'm not sure if its surprising that Hillsborough gets more than Caroni and Navet. That max value at Hollis looks pretty crazy.
# # Rolling Averages
# Plotting the rolling averages may give a better idea at how the rainfall patterns are changing. After some experimenting, I've found that a rolling window of 24 months gives the best representation.
roll_window = 24
df['Caroni_avg'] = df['Caroni'].rolling(window=roll_window).mean()
df['Navet_avg'] = df['Navet'].rolling(window=roll_window).mean()
df['Hollis_avg'] = df['Hollis'].rolling(window=roll_window).mean()
df['Hillsborough_avg'] = df['Hillsborough'].rolling(window=roll_window).mean()
df.head()
# # Data Visualization
C = df[['Caroni', 'Caroni_avg']].plot(figsize=(16,8), title='Caroni-Arena Reservoir Monthly Rainfall')
C.set_ylabel("Monthly Rainfall (mm)")
# At Caroni-Arena Reservoir, rainfall seems to be pretty constant. 2014 and 2015 in particular saw a lot of rainfall.
N = df[['Navet', 'Navet_avg']].plot(figsize=(16,8), title='Navet Reservoir Monthly Rainfall')
N.set_ylabel("Monthly Rainfall (mm)")
# At Navet Reservoir, no dominant pattern is present on the graph. Heavy rainfall in 1998 and 2001 skews the average upward, while low rainfall between 2014 and 2017 lowered the average.
Ho = df[['Hollis', 'Hollis_avg']].plot(figsize=(16,8), title='Hollis Reservoir Monthly Rainfall')
Ho.set_ylabel("Monthly Rainfall (mm)")
# At Hollis Reservoir, the appears to be constant. The regular fluctuations in the average line may be caused by the El Nino/ La Nina phenomenon. However after each cycle, the rainfall returns to the same level.
Hi = df[['Hillsborough', 'Hillsborough_avg']].plot(figsize=(16,8), title='Hillsborough Reservoir Monthly Rainfall')
Hi.set_ylabel("Monthly Rainfall (mm)")
# At Hillsborough reservoir in Tobago, the rainfall again has a cyclic pattern. From 2013 to 2017 there was a decrease in rainfall but is now looking to head up again, similar to what happend from 2008 to 2013.
# # Annual Rainfall Volumes
# Based on the monthly rainfall data, there is not enough evidence to suggest that rainfall is decreasing. Now we are going to plot the annual rainfall volumes to back up this claim. I'm going to create a new dataframe to hold these values.
annual_df = df[['Caroni', 'Navet', 'Hollis', 'Hillsborough']].groupby(df.index.year).sum() #groupby to organise in years
annual_df.head()
C = annual_df['Caroni'].plot(figsize=(16,8), title='Caroni-Arena Reservoir Annual Rainfall', grid=True)
C.set_ylabel("Annual Rainfall (mm)")
# The dip at the end is because all the values for 2018 are not in as yet. But as shown, there are no large deviations from the norm. I suspect this will be true for all of the reservoirs.
N = annual_df['Navet'].plot(figsize=(16,8), title='Navet Reservoir Annual Rainfall', grid=True)
N.set_ylabel("Annual Rainfall (mm)")
Ho = annual_df['Hollis'].plot(figsize=(16,8), title='Hollis Reservoir Annual Rainfall', grid=True)
Ho.set_ylabel("Annual Rainfall (mm)")
Hi = annual_df['Hillsborough'].plot(figsize=(16,8), title='Hillsborough Reservoir Annual Rainfall', grid=True)
Hi.set_ylabel("Annual Rainfall (mm)")
# Nothing suspicious at Hollis and Hillsborough. This reinforces the idea that there are no significant deviations from the normal rainfall patterns. Everybody is pretty close to that 2000 mm line, except for Hollis who is above.
# # Wet and Dry Season Variability
# The Dry season is traditionally considered to be January - May, and the wet season is June - December. Is there a shift in these periods? Are the seasons more distinguishable now or less?
# +
# Create a Seasonal Dictionary that will map months to seasons
SeasonDict = {11: 'Wet', 12: 'Wet', 1: 'Dry', 2: 'Dry', 3: 'Dry', 4: 'Dry', 5: 'Dry', 6: 'Wet', 7: 'Wet', 8: 'Wet', 9: 'Wet', 10: 'Wet'}
df['Season'] = ""
for row in df.index:
df.Season[row] = SeasonDict[row.month]
df.head()
# -
dry_df = df[['Caroni', 'Navet', 'Hollis', 'Hillsborough']][df.Season == 'Dry']
dry_df = dry_df.groupby(dry_df.index.year).sum() #groupby to organise in years
dry_df['Caroni_5year_avg'] = dry_df['Caroni'].rolling(window=5).mean()
dry_df['Navet_5year_avg'] = dry_df['Navet'].rolling(window=5).mean()
dry_df['Hollis_5year_avg'] = dry_df['Hollis'].rolling(window=5).mean()
dry_df['Hillsborough_5year_avg'] = dry_df['Hillsborough'].rolling(window=5).mean()
C = dry_df[['Caroni', 'Caroni_5year_avg']].plot(figsize=(16,8), title='Caroni-Arena Dry Season Rainfall', grid=True)
C.set_ylabel("Dry Season Rainfall (mm)")
C = dry_df[['Navet', 'Navet_5year_avg']].plot(figsize=(16,8), title='Navet Dry Season Rainfall', grid=True)
C.set_ylabel("Dry Season Rainfall (mm)")
C = dry_df[['Hollis', 'Hollis_5year_avg']].plot(figsize=(16,8), title='Hollis Dry Season Rainfall', grid=True)
C.set_ylabel("Dry Season Rainfall (mm)")
C = dry_df[['Hillsborough', 'Hillsborough_5year_avg']].plot(figsize=(16,8), title='Hillsborough Dry Season Rainfall', grid=True)
C.set_ylabel("Dry Season Rainfall (mm)")
# The plots of the dry season rainfall show no distinct patterns at the Caroni-Arena, Hollis, or Hillsborough Reservoirs. At Navet however, the rainfall appears to be decreasing. Now for the wet season.
wet_df = df[['Caroni', 'Navet', 'Hollis', 'Hillsborough']][df.Season == 'Wet']
wet_df = wet_df.groupby(wet_df.index.year).sum() #groupby to organise in years
wet_df['Caroni_5year_avg'] = wet_df['Caroni'].rolling(window=5).mean()
wet_df['Navet_5year_avg'] = wet_df['Navet'].rolling(window=5).mean()
wet_df['Hollis_5year_avg'] = wet_df['Hollis'].rolling(window=5).mean()
wet_df['Hillsborough_5year_avg'] = wet_df['Hillsborough'].rolling(window=5).mean()
C = wet_df[['Caroni', 'Caroni_5year_avg']].plot(figsize=(16,8), title='Caroni-Arena Wet Season Rainfall', grid=True)
C.set_ylabel("Wet Season Rainfall (mm)")
C = wet_df[['Navet', 'Navet_5year_avg']].plot(figsize=(16,8), title='Navet Wet Season Rainfall', grid=True)
C.set_ylabel("Wet Season Rainfall (mm)")
C = wet_df[['Hollis', 'Hollis_5year_avg']].plot(figsize=(16,8), title='Hollis Wet Season Rainfall', grid=True)
C.set_ylabel("Wet Season Rainfall (mm)")
C = wet_df[['Hillsborough', 'Hillsborough_5year_avg']].plot(figsize=(16,8), title='Hillsborough Wet Season Rainfall', grid=True)
C.set_ylabel("Wet Season Rainfall (mm)")
# Again, there is a decrease of rainfall at Navet Reservoir while the remainder of the reservoirs appear to be constant.
# # What about max and min values?
# Are those values deviating from the norm? It is possible that the average is not changing, but the max's are getting higher and the min's are getting lower. It doesn't look like that is the case from the graph, but let's plot some more lines just in case. I'm going to create yet another df. Minimum values are more concerning so those are first.
min_df = df[['Caroni', 'Navet', 'Hollis', 'Hillsborough']].groupby(df.index.year).min() #groupby to organise in years
min_df.head()
A = min_df.plot(figsize=(16,8), title='Minimum Monthly Rainfall', grid=True)
A.set_ylabel('Minimum Rainfall in a month by year (mm)')
# I've plotted all on the same graph because its all over the place and shows no patterns. Same for the maximum values below.
max_df = df[['Caroni', 'Navet', 'Hollis', 'Hillsborough']].groupby(df.index.year).max() #groupby to organise in years
max_df.head()
A = max_df.plot(figsize=(16,8), title='Maximum Monthly Rainfall', grid=True)
A.set_ylabel('Maximum Rainfall in a month by year (mm)')
# # Conclusions
# Based on this simple study, there is not enough evidence to suggest that rainfall at the major reservoirs across Trinidad and Tobago is decreasing. However in the dry season and wet plots at Navet reservoir, it was shown that rainfall has decreased since the 1990's, although this change is not obvious in the plots of the annual and monthly totals. It is recommended that this study be repeated with a larger dataset. Maybe it is possible that rainfall 50 years was more than what we receive today. Without data I can't backup that claim.
| 9,679 |
/link.ipynb | 42dd884fa7b22e030d28d242875ec537a4341e8a | [] | no_license | RuxuZhang/Meetup_Team_Project | https://github.com/RuxuZhang/Meetup_Team_Project | 0 | 0 | null | null | null | null | Jupyter Notebook | false | false | .py | 32,635 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
from sklearn import datasets
from sklearn.linear_model import LogisticRegression
import numpy as np
import matplotlib.pyplot as plt
iris = datasets.load_iris()
list(iris.keys())
#petal width
X = iris["data"][:,3:]
X[1:10]
# classify virginica against everything else
# Assign Class label: 1 if virginica , else 0
y = (iris["target"]==2).astype(np.int)
y
## Train logistic regression
log_reg = LogisticRegression()
model = log_reg.fit(X,y)
print("intercept(theta_0):", model.intercept_[0],"\nCoefficient(theta_1): ",model.coef_[0][0])
# Create a test set with petal widths between 0 and 3 cm
X_test = np.linspace(0,3,1000).reshape(-1,1)
X_test[1:5]
y_prob = log_reg.predict_proba(X_test)
plt.plot(X_test,y_prob[:,1],"g-",label="Virginica")
plt.plot(X_test,y_prob[:,0],"b--",label = "Not Virginica")
plt.axvline(x=1.61,color='r',linestyle="--",label="Decision boundary")
plt.xlabel("Petal width (cm)")
plt.ylabel("Probability")
plt.legend()
plt.show()
log_reg.predict([[1.7],[1.5]])
# Predict for petal width 1.7cm
p_17 = 1/(1+np.exp(4.22-(2.617*1.7)))
p_17
#Preict for petal width of 2.5cm
p_25 = 1/(1+np.exp(4.22-(2.617*2.5)))
p_25
topic_id == topic_a]
common_member_cnt = len(pd.merge(df_mem_topic[df_mem_topic.topic_id == topic_a], df_mem_topic[df_mem_topic.topic_id == topic_b], how='inner', on=['member_id']))
# print(common_member_cnt)
cor_val = common_member_cnt * common_member_cnt /(topic_member_lst[i] * topic_member_lst[j + i])
# print(cor_val)
print("_____END_____")
topic_corr[i][j + i] = cor_val
np.save("corr_result.npy", topic_corr)
| 1,879 |
/Basics -Regression- Classification-Fully Connected Network_Pytorch/Feed_Forward.ipynb | c904b73242bb80d8622c0c54c565ec8df526cefc | [] | no_license | prajinkhadka/Deep-Learning-With-Tensorflow-and-Pytorch | https://github.com/prajinkhadka/Deep-Learning-With-Tensorflow-and-Pytorch | 4 | 3 | null | null | null | null | Jupyter Notebook | false | false | .py | 702,127 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# +
import numpy as np
import matplotlib.pyplot as plt
import matplotlib.colors
import time
import pandas as pd
import math
# -
import torch
torch.manual_seed(0)
# +
# custom color mappings
my_cmap = matplotlib.colors.LinearSegmentedColormap("", ["red", "green", "yellow"])
# -
from sklearn.datasets import make_blobs
data, labels = make_blobs(n_samples=1000,centers =4, n_features=2, random_state=0)
print(data.shape, labels.shape)
plt.scatter(data[:,0], data[:,1], c=labels)
from sklearn.model_selection import train_test_split
# +
#splitting the data into train and test
X_train, X_val, Y_train, Y_val = train_test_split(data, labels, stratify=labels, random_state=0)
print(X_train.shape, X_val.shape, labels.shape)
# -
print(X_train.shape, Y_train.shape)
print(x_test.shape, y_test.shape)
# ## Feed Forward Network in Pytorch from scratch
# +
# converting the data into tensor
# -
#converting the numpy array to torch tensors
X_train, Y_train, X_val, Y_val = map(torch.tensor, (X_train, Y_train, X_val, Y_val))
print(X_train.shape, Y_train.shape)
# 
# +
# fucntion for creating the model
# vector notation
def model(x):
# a1 is the first layer, where inputs will be N and 2 is the number of features
# h1 is just the sigmoid
# a2 is the secodn alyers wherer inputs wull be N and 2 features
# a2 has 8 weights in total and after multiplying output would be (N,4)
a1 = torch.matmul(x, weights1)+ bias1 # (N ,2) * (2 ,2) -> (N,2)
h1 = a1.sigmoid() # (N,2)
a2 = torch.matmul(h1, weights2) + bias2 # (N,2)* ( 4, 4) -> (N ,4)
# -1 -> sum along the last dimension
# unsqueeze - add 1 in last dimension -> i.e N -> (N,1)
h2 = a2.exp()/ a2.exp().sum(-1).unsqueeze(-1) # (N ,4)
return h2
# +
# Now defining our cross entropy loss
# -
y_hat = torch.tensor([[0.1, 0.2, 0.3, 0.4], [0.8, 0.1, 0.05, 0.05]])
y= torch.tensor([2,0])
y
# here, y [2] , and y[0] are the ground truths.
#
# Now to compute the cross entropy loss
# # find element from y_hat given by y
#
#
# y_hat[range(y_hat.shape[0]), y].mean()
y_hat[range(y_hat.shape[0]), y].mean().item()
#function to calculate loss of a function.
#y_hat -> predicted & y -> actual
def loss_fn(y_hat, y):
return -(y_hat[range(y.shape[0]), y].log()).mean()
def accuracy(y_hat, y):
pred = torch.argmax(y_hat, dim=1)
return (pred ==y).float().mean()
plt.style.use("seaborn")
# # Training the model
# +
# set the seed
torch.manual_seed(0)
#initilaize the weights and bias -> He initialization
weights1 = torch.randn(2,2) / math.sqrt(2)
weights1.requires_grad_()
bias1 = torch.zeros(2, requires_grad= True)
weights2 = torch.randn(2,4) / math.sqrt(2)
weights2.requires_grad_()
bias2 = torch.zeros(4, requires_grad = True)
# +
# parameterts for the mode
# -
learning_rate = 0.2
epochs = 100
type(X_train)
# converting to float and long
X_train = X_train.float()
Y_train = Y_train.long()
X_val = X_val.float()
Y_val = Y_val.long()
# training th network
loss_arr =[]
acc_arr = []
val_acc_arr = []
for epoch in range(epochs):
y_hat = model(X_train) # computing the prediction
loss = loss_fn(y_hat, Y_train) # computing the loss
loss.backward()
loss_arr.append(loss.item())
acc_arr.append(accuracy(y_hat, Y_train))
with torch.no_grad():
val_acc_arr.append(accuracy(model(X_val), Y_val))
weights1 -= learning_rate * weights1.grad
weights2 -= learning_rate * weights2.grad
bias1 -= learning_rate * bias1.grad
bias2 -= learning_rate * bias2.grad
weights1.grad.zero_()
bias1.grad.zero_()
weights2.grad.zero_()
bias2.grad.zero_()
plt.plot(loss_arr, 'r-', label = 'loss')
plt.plot(acc_arr, 'b-', label ='accuracy')
plt.plot(val_acc_arr, 'g-', label='val_acc')
plt.grid()
plt.xlabel('epochs')
plt.legend()
# +
## Using nn.Functionl, ust replacing the loss wiht in built loss
import torch.nn.functional as F
# +
torch.manual_seed(0)
weights1 = torch.randn(2, 2) / math.sqrt(2)
weights1.requires_grad_()
bias1 = torch.zeros(2, requires_grad=True)
weights2 = torch.randn(2, 4) / math.sqrt(2)
weights2.requires_grad_()
bias2 = torch.zeros(4, requires_grad=True)
learning_rate = 0.2
epochs = 10000
loss_arr = []
acc_arr = []
for epoch in range(epochs):
y_hat = model(X_train) #compute the predicted distribution
loss = F.cross_entropy(y_hat, Y_train) #just replace the loss function with built in function
loss.backward()
loss_arr.append(loss.item())
acc_arr.append(accuracy(y_hat, Y_train))
with torch.no_grad():
weights1 -= weights1.grad * learning_rate
bias1 -= bias1.grad * learning_rate
weights2 -= weights2.grad * learning_rate
bias2 -= bias2.grad * learning_rate
weights1.grad.zero_()
bias1.grad.zero_()
weights2.grad.zero_()
bias2.grad.zero_()
plt.plot(loss_arr, 'r-', label='loss')
plt.plot(acc_arr, 'b-', label='train accuracy')
plt.legend(loc='best')
plt.title("Loss plot - nn.Functional")
plt.xlabel("Epoch")
plt.show()
plt.show()
print('Loss before training', loss_arr[0])
print('Loss after training', loss_arr[-1])
# -
# # Using nn.parameters
import torch.nn as nn
class FirstNetwork(nn.Module):
def __init__(self):
super().__init__()
torch.manual_seed(0)
#wrap all the weights and biases inside nn.parameter()
self.weights1 = nn.Parameter(torch.randn(2, 2) / math.sqrt(2))
self.bias1 = nn.Parameter(torch.zeros(2))
self.weights2 = nn.Parameter(torch.randn(2, 4) / math.sqrt(2))
self.bias2 = nn.Parameter(torch.zeros(4))
def forward(self, X):
a1 = torch.matmul(X, self.weights1) + self.bias1
h1 = a1.sigmoid()
a2 = torch.matmul(h1, self.weights2) + self.bias2
h2 = a2.exp()/a2.exp().sum(-1).unsqueeze(-1)
return h2
def fit(epochs = 10000, learning_rate = 0.2, title = ""):
loss_arr = []
acc_arr = []
for epoch in range(epochs):
y_hat = model(X_train) #forward pass
loss = F.cross_entropy(y_hat, Y_train) #loss calculation
loss_arr.append(loss.item())
acc_arr.append(accuracy(y_hat, Y_train))
loss.backward() #backpropagation
with torch.no_grad():
#updating the parameters
for param in model.parameters():
param -= learning_rate * param.grad
model.zero_grad() #setting the gradients to zero
plt.plot(loss_arr, 'r-', label='loss')
plt.plot(acc_arr, 'b-', label='train accuracy')
plt.legend(loc='best')
plt.title(title)
plt.xlabel("Epoch")
plt.show()
print('Loss before training', loss_arr[0])
print('Loss after training', loss_arr[-1])
# +
model= FirstNetwork()
fit(1000, 0.2)
# -
# ## Using NN.linear nad optim
# +
class FirstNetwork_v1(nn.Module):
def __init__(self):
super().__init__()
torch.manual_seed(0)
self.ln1 = nn.Linear(2,2)
self.ln2 = nn.Linear(2,4)
def forward(self,X):
a1 = self.ln1(X) # computes dot product and add the bias
h1 = a1.sigmoid()
a2 = self.ln2(h1)
h2 = a2.exp()/a2.exp().sum(-1).unsqueeze(-1)
return h2
# -
model_v1 = FirstNetwork_v1()
fit(10000, 0.2)
# #### using nn.optim instead of updating gradeints mansually
from torch import optim
def fit_v1(epochs=100,learning_rate = 0.2):
loss_accr =[]
acc_accr =[]
opt= optim.SGD(model.parameters(), lr = learning_rate)# optimzier
for epoch in range(epochs):
y_hat = model(X_train)
loss = F.cross_entropy(y_hat, Y_train)
loss_arr.append(loss.item())
acc_arr.append(accuracy(y_hat, Y_train))
loss.backward()
opt.step()
opt.zero_grad()
model = FirstNetwork_v1()
fit_v1(10000,0.2)
plt.plot(loss_arr, 'r-', label='loss')
plt.plot(acc_arr, 'b-', label='train accuracy')
plt.legend(loc='best')
plt.xlabel("Epoch")
plt.show()
print('Loss before training', loss_arr[0])
print('Loss after training', loss_arr[-1])
# # using nn.Sequential
class FirstNetwork_v2(nn.Module):
def __init__(self):
super().__init__()
torch.manual_seed(0)
self.net = nn.Sequential(
nn.Linear(2,2),
nn.Sigmoid(),
nn.Linear(2,4),
nn.Softmax())
def forward(self,X):
return self.net(X)
model = FirstNetwork_v2()
def fit_v2(x, y, model, opt, loss_fn, epochs =10000):
for epoch in range(epochs):
a = model(x)
loss = loss_fn(a , y)
loss.backward()
opt.step()
opt.zero_grad()
return loss.item()
# +
loss_fn = F.cross_entropy
opt = optim.SGD(model.parameters(), lr = 0.2)
fit_v2(X_train, Y_train, model, opt, loss_fn)
# -
# ## Diffferenct Weight Initialization
import torch
import torch.nn as nn
def init_weight(m):
if type(m) == nn.Linear:
torch.nn.init.xavier_uniform_(m.weight) # using uniform distributin
# torch.nn.init.kaimag_uniform(m.weight) # usinh heinitializton
m.bias.data.fill_(0)
net = nn.Sequential(nn.Linear(2,2), nn.Linear(2,4))
net
net.apply(init_weight)
# + active=""
# # Running on GPU
#
# device = torch.device("cuda") if torch.cuda.is_available() else torch.device("cpu") #create a device
# + active=""
# #moving inputs to GPU
# X_train=X_train.to(device)
# Y_train=Y_train.to(device)
#
# model = FirstNetwork_v2()
# model.to(device) #moving the network to GPU
#
# #calculate time
# tic = time.time()
# print('Final loss', fit_v2(X_train, Y_train, model, opt, loss_fn))
# toc = time.time()
# print('Time taken', toc - tic)
# -
| 10,122 |
/Keras_Regression IMP.ipynb | a17240fe3b8c7f3b9cc4382c55b6fe056da990f9 | [] | no_license | farooqahmed7/Keras- | https://github.com/farooqahmed7/Keras- | 0 | 0 | null | null | null | null | Jupyter Notebook | false | false | .py | 1,328,565 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Python [default]
# language: python
# name: python3
# ---
# # Landmanbot survey results
# library imports
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
# %matplotlib inline
from matplotlib.gridspec import GridSpec
# import file
df_raw = pd.read_excel('../surveyResults.xls', sheetname='Landmanbot')
df_raw.head(3)
# rename columns
df_edt = df_raw.rename(index=str, columns={"ID de la réponse": "id"
, "Date de soumission": "date"
, "Dernière page": "lastPage"
, "Langue de départ": "language"
, "Avez-vous utilisé Landmanbot -": "usedBot"
, "Avez-vous trouvé les informations que vous cherchiez -": "infoFound"
, "La qualité des réponses et du service vous a-t-elle convenu - []":"ansQuality"
, "Auriez-vous préféré une expérience plus libre ou plus guidée -":"moreGuided"
, "Aves-vous des remarques ou des suggestions d'amélioration -":"remarks"
, "Pourquoi n'avez-vous pas utilisé Landmanbot -":"whyNotUseBot"
, "Qu'est-ce qu'il manque pour que vous l'utilisiez -":"missingToUseBot"})
df_edt.head()
df_edt.info()
# drop rows with no datetime
df_edt.dropna(axis=0, how='any', thresh=None, subset=['date'], inplace=True)
df_edt.head()
len(df_edt)
# replace oui/non/NaN with 1/0/-999.25
df_edt['usedBot'].replace(regex=True,inplace=True,to_replace='Oui',value='1')
df_edt['usedBot'].replace(regex=True,inplace=True,to_replace='Non',value='0')
df_edt['usedBot'].replace(regex=True,inplace=True,to_replace=np.nan,value='-999.25')
df_edt['infoFound'].replace(regex=True,inplace=True,to_replace='Oui',value='1')
df_edt['infoFound'].replace(regex=True,inplace=True,to_replace='Non',value='0')
df_edt['infoFound'].replace(regex=True,inplace=True,to_replace=np.nan,value='-999.25')
df_edt.head()
df_edt.nunique()
# fractions of each value
#df_edt[df_edt['answQuality'] == 4]
fracs = pd.value_counts(df_edt['ansQuality'].values, sort=False)
fracs
# +
# pieplot of ansQuality
# Data to plot
labels = '1','2','4','5','6'
sizes = [1,2,3,8,5]
colors = ['gold', 'yellowgreen', 'lightcoral', 'lightskyblue', 'lightgrey']
explode = (0, 0, 0, 0, 0) # explode 1st slice
# Plot
plt.pie(sizes, explode=explode, labels=labels, colors=colors,
autopct='%1.1f%%', shadow=False, startangle=0)
plt.title('PiePlot of ansQuality')
plt.axis('equal')
plt.show()
# -
df_edt.head()
# nb of respondants who used bot
countUsedBot = len(df_edt[(df_edt['usedBot'] == '1')])
countUsedBot
# nb of respondants who found info
countInfoFound = len(df_edt[(df_edt['infoFound'] == '1')])
countInfoFound
# counts for each vote
countAnsQual_1 = len(df_edt[(df_edt['ansQuality'] == 1)])
countAnsQual_2 = len(df_edt[(df_edt['ansQuality'] == 2)])
countAnsQual_3 = len(df_edt[(df_edt['ansQuality'] == 3)])
countAnsQual_4 = len(df_edt[(df_edt['ansQuality'] == 4)])
countAnsQual_5 = len(df_edt[(df_edt['ansQuality'] == 5)])
countAnsQual_6 = len(df_edt[(df_edt['ansQuality'] == 6)])
inputList = countAnsQual_1,countAnsQual_2,countAnsQual_3,countAnsQual_4,countAnsQual_5,countAnsQual_6
inputList
# check total must = 19
countAnsQual_1+countAnsQual_2+countAnsQual_3+countAnsQual_4+countAnsQual_5+countAnsQual_6
# histogram of results
# results to nd.array
x = np.asarray([[1],[ 1, 1],[], [1, 1, 1], [1, 1, 1, 1, 1, 1, 1, 1], [1, 1, 1, 1, 1]])
ind = (1.025,1.05,1.075,1.1,1.125,1.150)
n, bins, patches = plt.hist(x, 6, facecolor='#2629e0')
plt.xlabel('Score attributed by respondant')
plt.xticks(ind, ('1','2','3','4','5','6'))
plt.ylabel('Count of scores attributed')
plt.title('Histogram of answer quality scores')
plt.axis([1.025, 1.15, 0, 6])
plt.grid(True)
plt.show()
df_edt.iloc[:10]
df_edt['moreGuided'][9]
plusGuide = len(df_edt[(df_edt['moreGuided'] == 'Plus guidée (Landmanbot me propose des messages/questions que je peux choisir)')])
plusLibre = len(df_edt[(df_edt['moreGuided'] == 'Plus libre (je peux parler à Landmanbot presque comme à un humain)')])
# +
# more guided vs more free
# pieplot of moreGuided
# Data to plot
labels = 'Plus libre','Plus guidée'
sizes = [plusLibre,plusGuide]
colors = ['#8ff442', '#f442eb']
explode = (0, 0)
# Plot
plt.pie(sizes, explode=explode, labels=labels, colors=colors,
autopct='%1.1f%%', startangle=90)
plt.title('Voulez-vous une experience plus libre ou plus guidée?')
plt.axis('equal')
plt.show()
# -
": 228} colab_type="code" id="UniqxGWdeH-y" outputId="b210c76d-3ead-4c71-b7fd-dcf31639bcd2"
df['date']
# + colab={} colab_type="code" id="QMYy8kvveL_Z"
df['date']=pd.to_datetime(df['date'])
# + colab={"base_uri": "https://localhost:8080/", "height": 228} colab_type="code" id="g_L5EfnpeWHy" outputId="6dc91ed7-5763-462e-be05-f4cd07e72eef"
df['date']
# + colab={} colab_type="code" id="CA2TndBveqx_"
def year_extraction(date):
return date.year
# + colab={} colab_type="code" id="K_T6lAaUebti"
df['year']=df['date'].apply(lambda date:date.year)# Similar to above function
df['month']=df['date'].apply(lambda date:date.month)
# + colab={"base_uri": "https://localhost:8080/", "height": 299} colab_type="code" id="z_knaKxGe5er" outputId="2d6299cd-7792-47c1-c6b4-5810122d11af"
df.head()
# + colab={"base_uri": "https://localhost:8080/", "height": 470} colab_type="code" id="L462LYAOfg42" outputId="6d2a3d0c-e5d9-4367-b208-3e9bba3a9c7c"
plt.figure(figsize=(12,8))
sb.boxplot(x='month',y='price',data=df)
# + colab={"base_uri": "https://localhost:8080/", "height": 263} colab_type="code" id="Ajwg0qacgRdf" outputId="838e9391-1863-436d-f12b-1bca4b4d8fc2"
df.groupby('month').mean()['price']
# + colab={} colab_type="code" id="nZdAOX0kgg_0"
df=df.drop('date',axis=1)
# + colab={"base_uri": "https://localhost:8080/", "height": 105} colab_type="code" id="pTvGd_AThR7p" outputId="ccd06a7f-7f1c-4101-dbb1-bc5132daee7e"
df.columns
# + colab={} colab_type="code" id="r1k2lrsVhb7k"
#df['zipcode'].value_counts()
# + colab={"base_uri": "https://localhost:8080/", "height": 215} colab_type="code" id="JU9403lEh6WY" outputId="29854b5c-9428-4261-e9a0-9fb5d9c94550"
df=df.drop('zipcode',axis=1)
df.head()
# + colab={"base_uri": "https://localhost:8080/", "height": 228} colab_type="code" id="C-h178_uiHOB" outputId="e5c647ed-df7b-4918-915e-214df288e949"
df['yr_renovated'].value_counts()
# + colab={"base_uri": "https://localhost:8080/", "height": 228} colab_type="code" id="CqL1Q-Zzi1Mp" outputId="f13afffa-00ad-4e42-e9ee-fee391f091c4"
df['sqft_basement'].value_counts()
# + colab={} colab_type="code" id="lO1tBjLXj7VJ"
X=df.drop('price',axis=1).values
y=df['price'].values
# + colab={} colab_type="code" id="m2k452r0p8qz"
from sklearn.model_selection import train_test_split
# + colab={} colab_type="code" id="feUHMcrep-6k"
X_train,X_test,y_train,y_test=train_test_split(X,y,test_size=0.3,random_state=101)
# + colab={} colab_type="code" id="wCy3N3GKqBZ8"
from sklearn.preprocessing import MinMaxScaler
# + colab={} colab_type="code" id="dGLqGeZDqE4Q"
scaler=MinMaxScaler()
# + colab={} colab_type="code" id="VdR5Lbm6qI25"
X_train=scaler.fit_transform(X_train)
# + colab={} colab_type="code" id="f6hf2Yl-qMG9"
X_test=scaler.transform(X_test)
# + colab={} colab_type="code" id="eK-bhDThqOsT"
from tensorflow.keras.models import Sequential
from tensorflow.keras.layers import Dense
# + colab={"base_uri": "https://localhost:8080/", "height": 34} colab_type="code" id="k1TA2IBVqwlJ" outputId="6b41c529-f496-446b-d2bf-d446a4f434e3"
X_train.shape
# + colab={} colab_type="code" id="bwZYNpbirRO9"
model=Sequential()
model.add(Dense(19,activation='relu'))
model.add(Dense(19,activation='relu'))
model.add(Dense(19,activation='relu'))
model.add(Dense(19,activation='relu'))
model.add(Dense(1))
model.compile(optimizer='adam',loss='mse')
# + colab={"base_uri": "https://localhost:8080/", "height": 1000} colab_type="code" id="7T2m1SSJryHI" outputId="f8ba36e2-2b1d-4220-a1bf-a1360aff14b6"
model.fit(X_train,y_train,
validation_data=(X_test,y_test),
batch_size=128,epochs=400)
# + colab={"base_uri": "https://localhost:8080/", "height": 1000} colab_type="code" id="8RyiW-Zcs6yU" outputId="35fa5752-0e66-4b0d-9d4f-43e87a478bf2"
model.history.history
# + colab={} colab_type="code" id="kmjUBOzOxg5a"
losses=pd.DataFrame(model.history.history)
# + colab={"base_uri": "https://localhost:8080/", "height": 294} colab_type="code" id="ox6JJAZpxo7b" outputId="74796da0-e883-42d4-8cb3-19c137ebba2f"
losses.plot()
# + colab={} colab_type="code" id="1T0QdyXrx2BL"
from sklearn.metrics import mean_squared_error,mean_absolute_error,explained_variance_score
# + colab={} colab_type="code" id="RAk1b_wYzmA6"
predictions=model.predict(X_test)
# + colab={"base_uri": "https://localhost:8080/", "height": 140} colab_type="code" id="yGutyGo10MnC" outputId="52e809ce-ca98-43b8-b40e-dc1867c49283"
predictions
# + colab={"base_uri": "https://localhost:8080/", "height": 34} colab_type="code" id="vSdSU2y70Odm" outputId="7ca551c3-b4dc-4152-9f3f-505f02e51323"
mean_squared_error(y_test,predictions)
# + colab={"base_uri": "https://localhost:8080/", "height": 34} colab_type="code" id="dV6cSW3E0WnF" outputId="7e7f9c2d-40a5-48c7-acbe-b079caedc222"
np.sqrt(mean_squared_error(y_test,predictions))
# + colab={"base_uri": "https://localhost:8080/", "height": 34} colab_type="code" id="f8Jnexyz0pJo" outputId="244f00da-a2b1-4f52-dae2-4d8229dde1a8"
mean_absolute_error(y_test,predictions)
# + colab={"base_uri": "https://localhost:8080/", "height": 175} colab_type="code" id="pJ6EX-Pu06mD" outputId="d69b7ae8-69fd-4605-a8e5-ffd44876321b"
df['price'].describe()
# + colab={"base_uri": "https://localhost:8080/", "height": 34} colab_type="code" id="jPPumTex1F31" outputId="969073e6-0b3c-468d-9309-4c69fff9c8ef"
5.402966e+05 # but mae is 88693, which is comparatively more
# + colab={"base_uri": "https://localhost:8080/", "height": 34} colab_type="code" id="nGJAg3s01Q44" outputId="0e785ace-d6c0-45ca-a7ad-dd262db11c20"
explained_variance_score(y_test,predictions)
# + colab={"base_uri": "https://localhost:8080/", "height": 306} colab_type="code" id="F-Lure_i1uSG" outputId="944437f5-57f2-4794-a2a8-4d6db35efcaf"
plt.scatter(y_test,predictions)
# + colab={"base_uri": "https://localhost:8080/", "height": 378} colab_type="code" id="q1izi3M-2DW7" outputId="37e7c648-43af-4a8b-c137-211ff547ee1e"
plt.figure(figsize=(12,6))
plt.scatter(y_test,predictions)
plt.plot(y_test,y_test,'r')
# + colab={} colab_type="code" id="qFtZfBLb2Y1g"
single_house=df.drop('price',axis=1).iloc[0]
# + colab={"base_uri": "https://localhost:8080/", "height": 105} colab_type="code" id="K6nnq89H2z5q" outputId="95dc605e-8e01-4276-95d8-36f3eee02db9"
single_house.values.reshape(-1,19)
# + colab={} colab_type="code" id="2DHDBkFB36Pl"
single_house=scaler.transform(single_house.values.reshape(-1,19))
# + colab={"base_uri": "https://localhost:8080/", "height": 34} colab_type="code" id="TTWdoE7I4I3R" outputId="43eeddb0-757c-4b39-cc09-a2e309d4c6fb"
model.predict(single_house)
# + colab={} colab_type="code" id="GZEFP4Mv4fTZ"
| 11,602 |
/Logistic_code_scratch.ipynb | e69c42a9d5547f944597c9655fc814f05f9ba666 | [] | no_license | aditis1204/Flight_Delay_Predictions | https://github.com/aditis1204/Flight_Delay_Predictions | 0 | 0 | null | null | null | null | Jupyter Notebook | false | false | .py | 17,987 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Python 2
# language: python
# name: python2
# ---
# ##### Test on synthetic data generated from the CRUST1.0 model
#
# This notebook runs a more complex test using synthetic data generated from the CRUST1.0 model. The area will be restricted to South America and the model will use a homogeneous density contrast along the anomalous Moho. Thus, we'll only the Moho depth information from CRUST1.0.
#
# The test is meant to simulate what we expect from the real data application ([south-america-moho.ipynb](south-america-moho.ipynb)). We will assume that the reference Moho depth and density contrast are unknown. Both of these hyperparameters, along with the regularization parameter, will be determined through cross-validation. For the reference depth and density contrast, we'll use seismic point data to score solutions. These data will also be simulated using the CRUST1.0 model.
# ## Package imports
#
# Load the necessary libraries to run the inversion and make graphs.
# Insert the plots into the notebook
# %matplotlib inline
# Load the standard scientific Python stack to numerical analysis and plotting.
# + jupyter={"outputs_hidden": false}
from __future__ import division, print_function
import multiprocessing
import zipfile
import datetime
import pickle
import itertools
import numpy as np
import matplotlib.pyplot as plt
from mpl_toolkits.basemap import Basemap
import seaborn # Makes the default style of the plots nicer
# -
# The computations generate a lot of run-time warnings. They aren't anything to be concerned about so disable them to avoid clutter.
# + jupyter={"outputs_hidden": true}
import warnings
warnings.simplefilter('ignore')
# -
# Load the required modules from [Fatiando a Terra](http://www.fatiando.org).
# + jupyter={"outputs_hidden": false}
from fatiando.vis import mpl
from fatiando.gravmag import tesseroid
from fatiando import gridder, utils
from fatiando.inversion import Smoothness2D
import fatiando
from fatiando.mesher import Tesseroid
# + jupyter={"outputs_hidden": false}
print("Version of Fatiando a Terra used: {}".format(fatiando.__version__))
# +
# pickle?
# -
# Load our custom classes and functions.
# + jupyter={"outputs_hidden": false}
from datasets import fetch_assumpcao_moho_points, fetch_crust1
from mohoinv import (MohoGravityInvSpherical, make_mesh, TesseroidRelief,
split_data, score_test_set, score_seismic_constraints,
score_all, fit_all, predict_seismic)
# -
# Get the number of cores in this computer to run the some things in parallel.
# + jupyter={"outputs_hidden": false}
ncpu = multiprocessing.cpu_count()
print("Number of cores: {}".format(ncpu))
# -
# ## Generate the synthetic data
# ### Load the CRUST1 model for South America
# Get the CRUST1.0 data from the archive file and cut it to South America.
# + jupyter={"outputs_hidden": false}
crust1 = fetch_crust1('../data/crust1.0.tar.gz').cut((-60, 20, -90, -30))
# -
# Establish a reference level for the Moho of the Normal Earth (given as height above the ellipsoid in meters, thus it is negative).
# + jupyter={"outputs_hidden": true}
reference = -30e3
# -
# Create a `TesseroidRelief` model of the Moho using a homogeneous density-contrast so that we can use in forward modeling.
# + jupyter={"outputs_hidden": true}
moho_density_contrast = 350
# + jupyter={"outputs_hidden": true}
model = TesseroidRelief(crust1.area, crust1.shape, relief=-crust1.moho_depth.ravel(),
reference=reference)
density = moho_density_contrast*np.ones(model.size)
density[model.relief < model.reference] *= -1
model.addprop('density', density)
# + jupyter={"outputs_hidden": false}
print('Tesseroids in the model (M_lat x M_lon): {} x {} = {}'.format(
model.shape[0], model.shape[1], model.size))
# -
# Make a plot of the Moho depth using an appropriate map projection.
# + jupyter={"outputs_hidden": false}
bm = Basemap(projection='cyl',
llcrnrlon=crust1.area[2], urcrnrlon=crust1.area[3],
llcrnrlat=crust1.area[0], urcrnrlat=crust1.area[1],
resolution='l')
# + jupyter={"outputs_hidden": false}
x, y = bm(model.lon.reshape(model.shape), model.lat.reshape(model.shape))
plt.figure(figsize=(7, 6))
plt.title('Moho depth')
bm.pcolormesh(x, y, -0.001*model.relief.reshape(model.shape), cmap='Greens')
plt.colorbar(pad=0.01).set_label('km')
bm.drawmeridians(np.arange(-80, -30, 10), labels=[0, 0, 0, 1], linewidth=0.2)
bm.drawparallels(np.arange(-50, 30, 15), labels=[1, 0, 0, 0], linewidth=0.2)
plt.tight_layout(pad=0)
# -
# ### Forward model the synthetic data
# Generate the computation grid for our synthetic dataset. The grid will have half the spacing of the model. This way, we'll have more points than we'll need to run the inversion. The extra points will be separated into a *test dataset* for cross-validation (see section [Cross-validation](#Cross-validation) below).
# + jupyter={"outputs_hidden": false}
# clon and clat are the coordinates of the center of each model cell
area = [model.clat.min(), model.clat.max(), model.clon.min(), model.clon.max()]
# Increase the shape to have half the grid spacing
full_shape = [s*2 - 1 for s in model.shape]
grid_height = 50e3
full_lat, full_lon, full_height = gridder.regular(area, full_shape, z=grid_height)
print('Number of grid points: {} x {} = {}'.format(full_shape[0], full_shape[1],
full_shape[0]*full_shape[1]))
print('Grid height: {} m'.format(grid_height))
# -
# Calculate the synthetic data and contaminate it with pseudo-random gaussian noise.
full_data_noisefree = tesseroid.gz(full_lon, full_lat, full_height, model, njobs=ncpu)
# + jupyter={"outputs_hidden": false}
#full_data = utils.contaminate(full_data_noisefree, 5, seed=0) # add unmodelled mass (mariani 2013 paper)
#look at top line of cell
intrusion = [Tesseroid(-55, -49, -27, -21, -30000, -45000, {"density":200})] #two notebooks one with intrusion one without
field = tesseroid.gz(full_lon, full_lat, full_height, intrusion)
full_data_noisefree += field
full_data = utils.contaminate(full_data_noisefree, 5, seed=0)
# + jupyter={"outputs_hidden": false}
x, y = bm(full_lon, full_lat)
ranges = np.abs([full_data.min(), full_data.max()]).max()
plt.figure(figsize=(7, 6))
plt.title('Moho synthetic gravity anomaly')
bm.contourf(x, y, full_data, 60, tri=True, cmap='RdBu_r', vmin=-ranges, vmax=ranges)
plt.colorbar(pad=0.01, aspect=50).set_label('mGal')
bm.drawmeridians(np.arange(-80, -30, 10), labels=[0, 0, 0, 1], linewidth=0.2)
bm.drawparallels(np.arange(-50, 30, 15), labels=[1, 0, 0, 0], linewidth=0.2)
plt.tight_layout()
# -
# ### Save the synthetic data and model
#
# We'll save the generated data and the corresponding model for later use and plotting. The data will be saved to the text file [../data/synthetic-data-crust1.txt](../data/synthetic-data-crust1.txt).
# + jupyter={"outputs_hidden": true}
now = datetime.datetime.utcnow().strftime('%d %B %Y %H:%M:%S UTC')
header = """# Generated by sinthetic-crust1.ipynb on {date}
# shape (nlat, nlon):
# {nlat} {nlon}
# lat lon height gravity_anomaly_noisefree gravity_anomaly_noisy
""".format(date=now, nlat=full_shape[0], nlon=full_shape[1])
with open('../data/synthetic-data-crust1.txt', 'w') as f:
f.write(header)
np.savetxt(f, np.c_[full_lat, full_lon, full_height, full_data_noisefree, full_data],
fmt='%.5f')
# -
# The model we'll save to a [Python pickle](https://docs.python.org/2/library/pickle.html) file. The `pickle` module allows us to serialize an object and load it back later. We'll use it to serialize the `model` object and save the file to the `model` folder.
# + jupyter={"outputs_hidden": true}
now = datetime.datetime.utcnow().strftime('%d %B %Y %H:%M:%S UTC')
model.metadata = "Generated by sinthetic-crust1.ipynb on {date}".format(date=now)
with open('../model/synthetic-crust1.pickle', 'w') as f:
pickle.dump(model, f)
# -
# ## Cross-validation
# First, we must separate the dataset into two parts: one for the inversion, another for cross-validation. The inversion dataset will have double the grid spacing and (for this test) must fall on top of each grid cell. The remaining data will be used for cross-validation.
# + jupyter={"outputs_hidden": false}
inversion_set, test_set, shape = split_data([full_lat, full_lon, full_height, full_data],
full_shape, every_other=2)
print("Number of inversion grid points: {} x {} = {}".format(shape[0], shape[1],
shape[0]*shape[1]))
print("Number of test set points: {}".format(test_set[0].size))
# -
# Test if the inversion set falls on top of the model cells.
# + jupyter={"outputs_hidden": true}
lat, lon, height, data = inversion_set
assert np.allclose(model.clon.ravel(), lon, rtol=1e-10, atol=0)
assert np.allclose(model.clat.ravel(), lat, rtol=1e-10, atol=0)
# -
# No errors means that both checks (`assert`) passed.
# Check if the score for the true model and error free data is 0 (perfect fit).
# + jupyter={"outputs_hidden": false}
score_test_set(model, full_lat, full_lon, full_height, full_data_noisefree, njobs=ncpu)
# -
# For this test, we'll assume that we **don't know the true reference level used or density contrast**. We'll try to estimate both through cross-validation. This means that there will be **two types of cross-validation**:
#
# 1. Testing agains the `test_set` for the regularization parameter
# 2. Testing against some other constraints for the reference level and density contrast
#
# These "other constraints" will come some point information about the Moho depth in some isolated points. In reality, these point depths could come from seismic experiments.
# We'll generate some fictional point constraints by taking the true value from our Moho model. The point coordinates chosen for this come from the seismic dataset of [Assumpção et al. (2013)](http://dx.doi.org/10.1016%2Fj.tecto.2012.11.014). We'll interpolate the values of the CRUST1 Moho depth onto these points.
# + jupyter={"outputs_hidden": true}
tmp = fetch_assumpcao_moho_points('../data/Moho_Map_SAm2013_data.tar.gz')
lat_points, lon_points = tmp[:2]
# + jupyter={"outputs_hidden": false}
seismic_points = gridder.interp_at(model.clat.ravel(), model.clon.ravel(),
model.relief, lat_points, lon_points)
# + jupyter={"outputs_hidden": false}
print('Number of seismic points: {}'.format(seismic_points.size))
# + jupyter={"outputs_hidden": true}
test_points = [lat_points, lon_points, seismic_points]
# + jupyter={"outputs_hidden": false}
x, y = bm(lon_points, lat_points)
plt.figure(figsize=(7, 6))
plt.title('Moho depth point constraints')
bm.scatter(x, y, c=-0.001*seismic_points, s=20, cmap='Greens')
plt.colorbar(pad=0.01, aspect=50).set_label('km')
bm.drawmeridians(np.arange(-80, -30, 10), labels=[0, 0, 0, 1], linewidth=0.2)
bm.drawparallels(np.arange(-50, 30, 15), labels=[1, 0, 0, 0], linewidth=0.2)
bm.drawcoastlines(color="#666666")
plt.tight_layout()
# -
# We can check if our scoring function from [mohoinv.py](mohoinv.py) works by feeding it the true model and the extracted points. The MSE should be zero.
# + jupyter={"outputs_hidden": false}
score_seismic_constraints(model, lat_points, lon_points, seismic_points)
# -
# Save our synthetic seismic constraints to a file so that we can plot it later.
# + jupyter={"outputs_hidden": true}
now = datetime.datetime.utcnow().strftime('%d %B %Y %H:%M:%S UTC')
header = """# Synthetic point estimates of Moho depth (from CRUST1.0)
# Generated by sinthetic-crust1.ipynb on {date}
# Moho depth is given in meters.
# lat lon depth
""".format(date=now)
with open('../data/crust1-point-depths.txt', 'w') as f:
f.write(header)
np.savetxt(f, np.c_[lat_points, lon_points, -seismic_points],
fmt='%.5f')
# -
# ## Inversion setup
#
# We need to generate a `TesseroidRelief` mesh for the inversion, give it a reference level and a density contrast. We'll assume that our mesh has the same geometry of the true model so that we can better compare the estimated and true Moho reliefs.
# + jupyter={"outputs_hidden": true}
mesh = model.copy(deep=True)
# -
# We'll also need an initial estimate, the solver and its configuration, and a regularization object.
# + jupyter={"outputs_hidden": false}
misfit = MohoGravityInvSpherical(lat, lon, height, data, mesh)
regul = Smoothness2D(mesh.shape)
initial = -60e3*np.ones(mesh.size)
# The initial estimate doesn't really matter too much
# -
# ## Plotting functions
#
# We'll define some plotting functions here to avoid having all this code down with the results. You can safely skip (not read) this section because we only define the functions here. They are called after the inversion below.
# + jupyter={"outputs_hidden": false}
def plot_fit(lat, lon, data, solution, bm):
ranges = np.abs([data.max(), data.min()]).max()
plt.figure(figsize=(7, 6))
plt.title('Observed (color) and predicted (contour) data')
levels = mpl.contourf(lon, lat, data, shape, 40, cmap='RdBu_r', basemap=bm,
vmin=-ranges, vmax=ranges)
plt.colorbar(pad=0.01).set_label('mGal')
mpl.contour(lon, lat, solution[0].predicted(), shape, levels,
basemap=bm, color='#333333')
bm.drawmeridians(np.arange(-80, -30, 10), labels=[0, 0, 0, 1], linewidth=0.2)
bm.drawparallels(np.arange(-50, 30, 15), labels=[1, 0, 0, 0], linewidth=0.2)
plt.tight_layout(pad=0)
def plot_residuals(solution):
residuals = solution[0].residuals()
plt.figure(figsize=(3, 2.5))
plt.text(0.65, 0.8,
"mean = {:.2f}\nstd = {:.2f}".format(residuals.mean(), residuals.std()),
transform=plt.gca().transAxes)
plt.hist(residuals, bins=20, normed=True, histtype='stepfilled')
plt.xlabel('Residuals (mGal)')
plt.ylabel('Normalized frequency')
plt.tight_layout(pad=0)
def plot_estimate(solution, bm):
moho = solution.estimate_
x, y = bm(moho.lons, moho.lats)
plt.figure(figsize=(7, 6))
plt.title("Estimated Moho depth")
bm.pcolormesh(x, y, -0.001*moho.relief.reshape(moho.shape), cmap='Greens')
plt.colorbar(pad=0.01).set_label('km')
bm.drawmeridians(np.arange(-80, -30, 10), labels=[0, 0, 0, 1], linewidth=0.2)
bm.drawparallels(np.arange(-50, 30, 15), labels=[1, 0, 0, 0], linewidth=0.2)
plt.tight_layout(pad=0)
def plot_diff(solution, model, bm):
moho = solution.estimate_
diff = -0.001*(model.relief - moho.relief).reshape(moho.shape)
ranges = np.abs([diff.max(), diff.min()]).max()
x, y = bm(moho.lons, moho.lats)
plt.figure(figsize=(7, 6))
plt.title("Difference between true and estimated")
bm.pcolormesh(x, y, diff, cmap='RdYlBu_r', vmin=-ranges, vmax=ranges)
plt.colorbar(pad=0.01).set_label('km')
bm.drawmeridians(np.arange(-40, 45, 20), labels=[0, 0, 0, 1], linewidth=0)
bm.drawparallels(np.arange(20, 65, 20), labels=[1, 0, 0, 0], linewidth=0)
plt.tight_layout(pad=0)
def plot_diff_hist(solution, model):
moho = solution.estimate_
diff = -0.001*(model.relief - moho.relief)
plt.figure(figsize=(3, 2.5))
plt.title('Difference (true model)')
plt.text(0.65, 0.8,
"mean = {:.2f}\nstd = {:.2f}".format(diff.mean(), diff.std()),
transform=plt.gca().transAxes)
# Use the line above so the text coordinates are in axes coordinates (0 to 1)
# instead of data coordinates, which may vary between runs.
plt.hist(diff.ravel(), bins=20, normed=True, histtype='stepfilled')
plt.xlabel('Differences (km)')
plt.ylabel('Normalized frequency')
plt.tight_layout(pad=0)
def plot_diff_seismic(solution, test_points, bm):
moho = solution.estimate_
x, y = bm(moho.lons, moho.lats)
diff = -0.001*(test_points[-1] - predict_seismic(moho, *test_points[:2]))
ranges = np.abs([diff.max(), diff.min()]).max()
lat, lon, depth = test_points
xp, yp = bm(lon, lat)
fig = plt.figure(figsize=(7, 6))
bm.pcolormesh(x, y, -0.001*moho.relief.reshape(moho.shape), cmap='Greens')
plt.colorbar(pad=0.01, aspect=50).set_label('Estimated Moho depth (km)')
bm.scatter(xp, yp, c=diff, s=40, cmap='PuOr_r',
vmin=-ranges, vmax=ranges, linewidths=0.1)
cb = plt.colorbar(pad=0.01, aspect=50)
cb.set_label('Difference between estimated and seismic (km)')
bm.drawmeridians(np.arange(-80, -30, 10), labels=[0, 0, 1, 0], linewidth=0.2)
bm.drawparallels(np.arange(-50, 30, 15), labels=[1, 0, 0, 0], linewidth=0.2)
plt.tight_layout(pad=0)
def plot_diff_seismic_hist(solution, test_points):
moho = solution.estimate_
diff = -0.001*(test_points[-1] - predict_seismic(moho, *test_points[:2]))
plt.figure(figsize=(3, 2.5))
plt.title('Difference (seismic points)')
plt.text(0.65, 0.8,
"mean = {:.2f}\nstd = {:.2f}".format(diff.mean(), diff.std()),
transform=plt.gca().transAxes)
# Use the line above so the text coordinates are in axes coordinates (0 to 1)
# instead of data coordinates, which may vary between runs.
plt.hist(diff, bins=20, normed=True, histtype='stepfilled')
plt.xlabel('Differences (km)')
plt.ylabel('Normalized frequency')
plt.tight_layout(pad=0)
def plot_cv_regul(regul_params, scores, best, log=True):
plt.figure(figsize=(5, 3))
plt.title('Cross-validation (regularization parameter)')
plt.plot(regul_params, scores, marker='o')
plt.plot(regul_params[best], scores[best], 's', markersize=10,
color=seaborn.color_palette()[2], label='Minimum')
plt.legend(loc='upper left')
plt.xscale('log')
if log:
plt.yscale('log')
plt.xlabel('Regularization parameter')
plt.ylabel(u'Mean Square Error')
plt.tight_layout(pad=0)
def plot_cv_ref_dens(densities, reference_levels, scores, best_dens, best_ref):
plt.figure(figsize=(5, 3))
plt.title('Cross-validation (reference level and density)')
plt.contourf(-0.001*reference_levels, densities, scores, 30, cmap='BuPu_r')
plt.colorbar(pad=0.01).set_label('Mean Square Error')
plt.plot(-0.001*reference_levels[best_ref], densities[best_dens], 's', markersize=10,
color=seaborn.color_palette()[2], label='Minimum')
l = plt.legend(loc='upper left')
for txt in l.get_texts():
txt.set_color('#ffffff')
plt.xlabel('Reference level (km)')
plt.ylabel(u'Density contrast (kg/m³)')
plt.tight_layout(pad=0)
def plot_convergence(solution, log=True):
plt.figure(figsize=(5, 3))
plt.title('Convergence')
plt.plot(range(solution.stats_['iterations'] + 1), solution.stats_['objective'])
plt.xlabel('Iteration')
plt.ylabel('Goal function')
if log:
plt.yscale('log')
plt.tight_layout(pad=0)
# -
# ## Run the inversion and cross-validations
# We'll keep the results in a Python dictionary (`dict`) along with all configuration and other metadata. We can then save this dict to a Pickle file and have inversion information saved with the results.
# + jupyter={"outputs_hidden": true}
results = dict()
# -
# Save also the configuration for the solver. We'll use the Gauss-Newton formulation of the inversion.
# + jupyter={"outputs_hidden": true}
results['config'] = dict(method='newton', initial=initial, tol=0.2, maxit=15)
# -
# These are the values of the regularization parameter, reference level, and density contrast that we'll use.
# + jupyter={"outputs_hidden": false}
results['regul_params'] = np.logspace(-5, -3, 3)
results['regul_params']
# + jupyter={"outputs_hidden": false}
results['reference_levels'] = np.arange(-32.5e3, -27.5e3 + 1, 2500)
results['reference_levels']
# + jupyter={"outputs_hidden": false}
results['densities'] = np.arange(300, 400 + 1, 50)
results['densities']
# -
# First, run the cross-validation to find the regularization parameter. We'll use one of the values for the reference and density contrast. The value of the regularization parameter that we estimate here will be used in the second cross-validation to find the density contrast and reference level.
# + jupyter={"outputs_hidden": false}
misfit.set_density(results['densities'][-1]).set_reference(results['reference_levels'][-1])
# -
# Run the inversion for each value in `regul_params` (in parallel using all available cores).
# + jupyter={"outputs_hidden": false}
solvers = [(misfit + mu*regul).config(**results['config'])
for mu in results['regul_params']]
# %time solutions = fit_all(solvers, njobs=ncpu)
# -
# Keep only the estimated models in the results dict instead of the whole solvers to reduce the size of the pickle file. We can calculate the predicted data, etc, from the model only.
# + jupyter={"outputs_hidden": true}
results['models_regul'] = [s.estimate_ for s in solutions]
# -
# Score the results against the test dataset.
# + jupyter={"outputs_hidden": false}
# %%time
results['scores_regul'] = score_all(results['models_regul'], test_set,
points=False, njobs=ncpu)
# -
# The best solution is the one with the smallest cross-validation score.
# + jupyter={"outputs_hidden": true}
best_regul = np.argmin(results['scores_regul'])
results['best_regul'] = best_regul
# -
# We'll use this solution as the inversion solver for the next cross-validation (for the reference level and density).
# + jupyter={"outputs_hidden": true}
results['solution_regul'] = solutions[best_regul]
# -
# But first, let's take a look at the current solution.
#
# Plot the cross-validation scores.
# + jupyter={"outputs_hidden": false}
plot_cv_regul(results['regul_params'], results['scores_regul'],
results['best_regul'], log=True)
plt.grid(True, which='both', axis='y')
# -
# The inversion residuals (to see if the solution fits the data).
# + jupyter={"outputs_hidden": false}
plot_residuals(results['solution_regul'])
# -
# The solution itself to see if it is smooth enough.
# + jupyter={"outputs_hidden": false}
plot_estimate(results['solution_regul'], bm)
# -
# Now, run the second cross-validation to estimate the density-contrast and the reference level.
# + jupyter={"outputs_hidden": true}
def set_ref_dens(solver, ref, dens):
"""
Configure the solver to use this reference level and density.
"""
res = solver.copy(deep=True)
# res is a multi-objective with the misfit function + regularization
# res[0] is the misfit (our inversion class)
# res[1] is the Smoothness2D instance
res[0].set_density(dens).set_reference(ref)
return res
# + jupyter={"outputs_hidden": false}
solvers = [set_ref_dens(results['solution_regul'], ref, dens)
for dens in results['densities']
for ref in results['reference_levels']]
# %time solutions = fit_all(solvers, njobs=ncpu)
# -
# Keep the only the estimated models in the results dict.
# + jupyter={"outputs_hidden": true}
results['models_refdens'] = [s.estimate_ for s in solutions]
# -
# Score the estimates against the seismic constraints.
# + jupyter={"outputs_hidden": true}
cv_shape = (len(results['densities']), len(results['reference_levels']))
# -
# Putting this step into a for loop
np.random.seed(1)
# +
# %%time
# for loop set array of certain size e.g. 625 and put into set_train definition
# after score is calculated need to put into array to store all values
train_size = np.array([625, 703, 750]) # for loop in for loop. loop with a bunch of different training sizes
iteration_number = 100
MSE_values = np.empty([train_size.size, iteration_number])
for j in range(train_size.size):
for i in range(iteration_number):
indices = np.arange(lat_points.size)
set_train = np.random.choice(indices, size=train_size[j], replace=False)
lat_train = lat_points[set_train]
lon_train = lon_points[set_train]
seis_train = seismic_points[set_train]
train_points = [lat_train, lon_train, seis_train]
lat_test = np.delete(lat_points, set_train)
lon_test = np.delete(lon_points, set_train)
seismic_test = np.delete(seismic_points, set_train)
results['scores_refdens'] = score_all(results['models_refdens'], train_points,
points=True, njobs=ncpu).reshape(cv_shape)
best = np.nanargmin(results['scores_refdens'])
results['solution'] = solutions[best]
# Find the index in reference_levels and densities corresponding to best
MSE_values[j, i] = score_seismic_constraints(results['solution'].estimate_, lat_test, lon_test, seismic_test)
#print(MSE_values) #np.save function to save results
np.save('MSE_values.npy',MSE_values)
# after done put this code into south american moho
# before plotting and saving data files use np.sqrt on MSE to get root mean square error
# -
np.save('MSE_values_625.npy',MSE_values[0, :])
np.save('MSE_values_703.npy',MSE_values[1, :])
np.save('MSE_values_750.npy',MSE_values[2, :])
# +
#np.isnan(MSE_values) #np.any
# +
#plotting MSE for training size 625 or about 2/3 of data
plt.figure()
plt.title("A histogram of Root Mean Square Errors from cross validation, training size = 625")
plt.hist(np.sqrt(MSE_values[0, :]))
plt.xlabel("RMSE in meters")
plt.ylabel("Frequency")
plt.text(2650, 19, "mean = {:.0f}\nstd = {:.1f}".format(np.mean(np.sqrt(MSE_values[0, :])), np.std(np.sqrt(MSE_values[0, :])))) #{:.0f}
plt.axvline(np.mean(np.sqrt(MSE_values[0, :])), color='k', linestyle='dashed')
plt.savefig("RMSE_training_size_625.jpg", dpi=100, bbox_inches="tight")
#plotting MSE for training size 703 or about 3/4 of data
plt.figure()
plt.title("A histogram of Root Mean Square Errors from cross validation, training size = 703")
plt.hist(np.sqrt(MSE_values[1, :]))
plt.xlabel("RMSE in meters")
plt.ylabel("Frequency")
plt.text(2750, 16, "mean = {:.0f}\nstd = {:.1f}".format(np.mean(np.sqrt(MSE_values[1, :])), np.std(np.sqrt(MSE_values[1, :]))))
plt.axvline(np.mean(np.sqrt(MSE_values[1, :])), color='k', linestyle='dashed')
plt.savefig("RMSE_training_size_703.jpg", dpi=100, bbox_inches="tight")
#plotting MSE for training size 750 or about 4/5 of data
plt.figure()
plt.title("A histogram of Root Mean Square Errors from cross validation, training size = 750")
plt.hist(np.sqrt(MSE_values[2, :]))
plt.xlabel("RMSE in meters")
plt.ylabel("Frequency")
plt.text(2780, 21, "mean = {:.0f}\nstd = {:.1f}".format(np.mean(np.sqrt(MSE_values[2, :])), np.std(np.sqrt(MSE_values[2, :]))))
plt.axvline(np.mean(np.sqrt(MSE_values[2, :])), color='k', linestyle='dashed')
plt.savefig("RMSE_training_size_750.jpg", dpi=100, bbox_inches="tight")
# -
results['solution'] = solutions[best]
# Find the index in reference_levels and densities corresponding to best
results['best_dens'], results['best_ref'] = np.unravel_index(best, cv_shape)
# Print the estimated parameters:
# + jupyter={"outputs_hidden": false}
estimated_ref = results['reference_levels'][results['best_ref']]
estimated_dens = results['densities'][results['best_dens']]
estimated_regul = results['regul_params'][results['best_regul']]
print('Cross-validation results:')
print(u' reference level: {} km (true = {})'.format(
-0.001*estimated_ref, -0.001*reference))
print(u' density contrast: {} kg/m³ (true = {})'.format(
estimated_dens, moho_density_contrast))
print(u' regularization parameter: {}'.format(estimated_regul))
# -
# Plot the cross-validation score for the reference level and density-contrast on map.
# + jupyter={"outputs_hidden": false}
plot_cv_ref_dens(results['densities'], results['reference_levels'],
results['scores_refdens'], results['best_dens'], results['best_ref'])
# -
# ### Save the results to a pickle file
# All of the above takes some time to run. We can now save the `results` dictionary to a pickle file and load it later to make plots of the results without re-calculating. Since the resulting pickle file will be large, we'll store it in a `zip` archive.
# + jupyter={"outputs_hidden": false}
# Dump the results dict to a pickle file
now = datetime.datetime.utcnow().strftime('%d %B %Y %H:%M:%S UTC')
results['metadata'] = "Generated by sinthetic-crust1.ipynb on {date}".format(date=now)
fname = 'synthetic-crust1'
pickle_file = '{}.pickle'.format(fname)
with open('results/{}'.format(pickle_file), 'w') as f:
pickle.dump(results, f)
# Zip the pickle file
zipargs = dict(mode='w', compression=zipfile.ZIP_DEFLATED)
with zipfile.ZipFile('results/{}.zip'.format(fname), **zipargs) as f:
f.write('results/{}'.format(pickle_file), arcname=pickle_file)
# -
# ### Plot the final solution
# + jupyter={"outputs_hidden": false}
plot_fit(lat, lon, data, results['solution'], bm)
# + jupyter={"outputs_hidden": false}
plot_residuals(results['solution'])
# + jupyter={"outputs_hidden": false}
plot_estimate(results['solution'], bm)
# + jupyter={"outputs_hidden": false}
plot_diff(results['solution'], model, bm)
# + jupyter={"outputs_hidden": false}
plot_diff_hist(results['solution'], model)
# + jupyter={"outputs_hidden": false}
plot_diff_seismic(results['solution'], test_points, bm)
# + jupyter={"outputs_hidden": false}
plot_diff_seismic_hist(results['solution'], test_points)
# -
np.sqrt(score_seismic_constraints(results['solution'].estimate_, lat_points, lon_points, seismic_points))
| 29,701 |
/notebooks/dconv.ipynb | e7e61d30fbc7247a5c421cedb286a1d7bd43e784 | [] | no_license | 395t/coding-assignment-week-10-rec-2 | https://github.com/395t/coding-assignment-week-10-rec-2 | 0 | 0 | null | null | null | null | Jupyter Notebook | false | false | .py | 2,080,650 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Python 3
# name: python3
# ---
# + [markdown] id="view-in-github" colab_type="text"
# <a href="https://colab.research.google.com/github/ValerieLangat/.../blob/master/Valerie_Langat_LS_DS_112_Loading_Data.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# + [markdown] id="-c0vWATuQ_Dn" colab_type="text"
# # Lambda School Data Science - Loading, Cleaning and Visualizing Data
#
# Objectives for today:
# - Load data from multiple sources into a Python notebook
# - !curl method
# - CSV upload method
# - Create basic plots appropriate for different data types
# - Scatter Plot
# - Histogram
# - Density Plot
# - Pairplot
# - "Clean" a dataset using common Python libraries
# - Removing NaN values "Interpolation"
# + [markdown] id="grUNOP8RwWWt" colab_type="text"
# # Part 1 - Loading Data
#
# Data comes in many shapes and sizes - we'll start by loading tabular data, usually in csv format.
#
# Data set sources:
#
# - https://archive.ics.uci.edu/ml/datasets.html
# - https://github.com/awesomedata/awesome-public-datasets
# - https://registry.opendata.aws/ (beyond scope for now, but good to be aware of)
#
# Let's start with an example - [data about flags](https://archive.ics.uci.edu/ml/datasets/Flags).
# + [markdown] id="wxxBTeHUYs5a" colab_type="text"
# ## Lecture example - flag data
# + id="nc-iamjyRWwe" colab_type="code" outputId="603b4d26-9aaa-4c0c-8003-d54e02e07bce" colab={"base_uri": "https://localhost:8080/", "height": 3315}
# Step 1 - find the actual file to download
# From navigating the page, clicking "Data Folder"
flag_data_url = 'https://archive.ics.uci.edu/ml/machine-learning-databases/flags/flag.data'
# You can "shell out" in a notebook for more powerful tools
# https://jakevdp.github.io/PythonDataScienceHandbook/01.05-ipython-and-shell-commands.html
# Funny extension, but on inspection looks like a csv
# !curl https://archive.ics.uci.edu/ml/machine-learning-databases/flags/flag.data
# Extensions are just a norm! You have to inspect to be sure what something is
# + id="UKfOq1tlUvbZ" colab_type="code" colab={}
# Step 2 - load the data
# How to deal with a csv? 🐼
import pandas as pd
flag_data = pd.read_csv(flag_data_url)
# + id="exKPtcJyUyCX" colab_type="code" outputId="b490d0f2-8c0e-45fd-86c6-32475f203ec7" colab={"base_uri": "https://localhost:8080/", "height": 233}
# Step 3 - verify we've got *something*
flag_data.head()
# + id="rNmkv2g8VfAm" colab_type="code" outputId="666c7c9b-dae9-4f9c-c344-1dab6b9d56bc" colab={"base_uri": "https://localhost:8080/", "height": 544}
# Step 4 - Looks a bit odd - verify that it is what we want
flag_data.count()
# + id="iqPEwx3aWBDR" colab_type="code" outputId="6f7fe2c9-18e9-473a-ba82-f0a296522c31" colab={"base_uri": "https://localhost:8080/", "height": 85}
# !curl https://archive.ics.uci.edu/ml/machine-learning-databases/flags/flag.data | wc
# + id="5R1d1Ka2WHAY" colab_type="code" outputId="08744ce2-5861-40d9-e47d-76764f3ae2dc" colab={"base_uri": "https://localhost:8080/", "height": 4984}
# So we have 193 observations with funny names, file has 194 rows
# Looks like the file has no header row, but read_csv assumes it does
help(pd.read_csv)
# + id="EiNiR6vExQUt" colab_type="code" colab={}
# ?pd.read_csv
# + id="oQP_BuKExQWE" colab_type="code" colab={}
??pd.read_csv
# + id="o-thnccIWTvc" colab_type="code" outputId="7cf1dab7-8484-4ddb-f83e-25783ab141a7" colab={"base_uri": "https://localhost:8080/", "height": 233}
# Alright, we can pass header=None to fix this
flag_data = pd.read_csv(flag_data_url, header=None)
flag_data.head()
# + id="iG9ZOkSMWZ6D" colab_type="code" outputId="7076d46e-a726-4e29-8434-bd8f8a43edbf" colab={"base_uri": "https://localhost:8080/", "height": 544}
flag_data.count()
# + id="gMcxnWbkWla1" colab_type="code" outputId="b4f6bb14-d735-4599-a0b9-e1cee23b9632" colab={"base_uri": "https://localhost:8080/", "height": 544}
flag_data.isna().sum()
# + [markdown] id="AihdUkaDT8We" colab_type="text"
# ### Yes, but what does it *mean*?
#
# This data is fairly nice - it was "donated" and is already "clean" (no missing values). But there are no variable names - so we have to look at the codebook (also from the site).
#
# ```
# 1. name: Name of the country concerned
# 2. landmass: 1=N.America, 2=S.America, 3=Europe, 4=Africa, 4=Asia, 6=Oceania
# 3. zone: Geographic quadrant, based on Greenwich and the Equator; 1=NE, 2=SE, 3=SW, 4=NW
# 4. area: in thousands of square km
# 5. population: in round millions
# 6. language: 1=English, 2=Spanish, 3=French, 4=German, 5=Slavic, 6=Other Indo-European, 7=Chinese, 8=Arabic, 9=Japanese/Turkish/Finnish/Magyar, 10=Others
# 7. religion: 0=Catholic, 1=Other Christian, 2=Muslim, 3=Buddhist, 4=Hindu, 5=Ethnic, 6=Marxist, 7=Others
# 8. bars: Number of vertical bars in the flag
# 9. stripes: Number of horizontal stripes in the flag
# 10. colours: Number of different colours in the flag
# 11. red: 0 if red absent, 1 if red present in the flag
# 12. green: same for green
# 13. blue: same for blue
# 14. gold: same for gold (also yellow)
# 15. white: same for white
# 16. black: same for black
# 17. orange: same for orange (also brown)
# 18. mainhue: predominant colour in the flag (tie-breaks decided by taking the topmost hue, if that fails then the most central hue, and if that fails the leftmost hue)
# 19. circles: Number of circles in the flag
# 20. crosses: Number of (upright) crosses
# 21. saltires: Number of diagonal crosses
# 22. quarters: Number of quartered sections
# 23. sunstars: Number of sun or star symbols
# 24. crescent: 1 if a crescent moon symbol present, else 0
# 25. triangle: 1 if any triangles present, 0 otherwise
# 26. icon: 1 if an inanimate image present (e.g., a boat), otherwise 0
# 27. animate: 1 if an animate image (e.g., an eagle, a tree, a human hand) present, 0 otherwise
# 28. text: 1 if any letters or writing on the flag (e.g., a motto or slogan), 0 otherwise
# 29. topleft: colour in the top-left corner (moving right to decide tie-breaks)
# 30. botright: Colour in the bottom-left corner (moving left to decide tie-breaks)
# ```
#
# Exercise - read the help for `read_csv` and figure out how to load the data with the above variable names. One pitfall to note - with `header=None` pandas generated variable names starting from 0, but the above list starts from 1...
# + id="okEjAUHwEZtE" colab_type="code" colab={}
# + [markdown] id="XUgOnmc_0kCL" colab_type="text"
# ## Loading from a local CSV to Google Colab
# + id="-4LA4cNO0ofq" colab_type="code" colab={}
# + [markdown] id="aI2oN4kj1uVQ" colab_type="text"
# # Part 2 - Basic Visualizations
# + [markdown] id="INqBGKRl88YD" colab_type="text"
# ## Basic Data Visualizations Using Matplotlib
# + id="r5_la-d52GcG" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 204} outputId="cafe4006-d2f1-458e-fa54-a795e258246a"
diabetes.head()
# + id="6FsdkKuh8_Rz" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 295} outputId="122565d2-6e3c-4135-bf9d-f845f4ce28f8"
import matplotlib.pyplot as plt
diabetes = pd.read_csv('https://raw.githubusercontent.com/ryanleeallred/datasets/master/diabetes.csv')
# Scatter Plot
plt.scatter(diabetes.Pregnancies, diabetes.BloodPressure)
plt.title('Pregnancies and Blood Pressure')
plt.xlabel('Pregnancies')
plt.ylabel('Blood Pressure')
plt.show()
# + id="huwUQ7zE9gkD" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 300} outputId="d5e81104-8e84-4e3d-94a5-4ff616de80e4"
# Histogram
plt.hist(diabetes['Pregnancies'], bins=10)
plt.xlabel('Pregnancies')
# + id="CSmpwXQN9o8o" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 300} outputId="0d2b9e09-83a7-4441-fae3-6dc7ece4dd8b"
# Seaborn Density Plot
import seaborn as sns
sns.distplot(diabetes.Pregnancies, hist=False)
# + id="TMMJG5rQ-g_8" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 1649} outputId="0ef6032f-dc3b-4841-e21f-3d7bfa00e600"
# Seaborn Pairplot
graph = sns.pairplot(diabetes)
# + [markdown] id="ipBQKbrl76gE" colab_type="text"
# ## Create the same basic Visualizations using Pandas
# + id="qWIO8zuhArEr" colab_type="code" colab={}
# Pandas Histogram - Look familiar?
# + id="zxEajNvjAvfB" colab_type="code" colab={}
# Pandas Scatterplot
# + id="XjR5i6A5A-kp" colab_type="code" colab={}
# Pandas Scatter Matrix - Usually doesn't look too great.
# + [markdown] id="tmJSfyXJ1x6f" colab_type="text"
# # Part 3 - Deal with Missing Values
# + [markdown] id="bH46YMHEDzpD" colab_type="text"
# ## Diagnose Missing Values
#
# Lets use the Adult Dataset from UCI. <https://github.com/ryanleeallred/datasets>
# + id="NyeZPpxRD1BA" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 306} outputId="be351aa7-aad3-455b-85ac-23a69d23e31d"
df_1 = pd.read_csv('https://raw.githubusercontent.com/ryanleeallred/datasets/master/adult.csv')
df_1.head()
# + [markdown] id="SYK5vXqt7zp1" colab_type="text"
# ## Fill Missing Values
# + id="32ltklnQ71A6" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 289} outputId="20b004a7-c631-4e19-df8c-fc3e94ed865f"
df_1.isnull().sum()
# + id="fJjtua128Ux1" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 253} outputId="eef37090-761d-47a9-a237-4bc08dc5e56d"
df_1 = pd.read_csv('https://raw.githubusercontent.com/ryanleeallred/datasets/master/adult.csv', na_values='Bachelors')
df.head()
# + [markdown] id="nPbUK_cLY15U" colab_type="text"
# ## Your assignment - pick a dataset and do something like the above
#
# This is purposely open-ended - you can pick any data set you wish. It is highly advised you pick a dataset from UCI or a similar semi-clean source. You don't want the data that you're working with for this assignment to have any bigger issues than maybe not having headers or including missing values, etc.
#
# After you have chosen your dataset, do the following:
#
# - Import the dataset using the method that you are least comfortable with (!curl or CSV upload).
# - Make sure that your dataset has the number of rows and columns that you expect.
# - Make sure that your dataset has appropriate column names, rename them if necessary.
# - If your dataset uses markers like "?" to indicate missing values, replace them with NaNs during import.
# - Identify and fill missing values in your dataset (if any)
# - Don't worry about using methods more advanced than the `.fillna()` function for today.
# - Create one of each of the following plots using your dataset
# - Scatterplot
# - Histogram
# - Density Plot
# - Pairplot (note that pairplots will take a long time to load with large datasets or datasets with many columns)
#
# If you get that done and want to try more challenging or exotic things, go for it! Use documentation as illustrated above, and follow the 20-minute rule (that is - ask for help if you're stuck!).
#
# If you have loaded a few traditional datasets, see the following section for suggested stretch goals.
# + id="NJdISe69ZT7E" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 253} outputId="7d9966f6-d376-4c81-f425-3c1f4c2dcec3"
df = pd.read_csv('https://raw.githubusercontent.com/ValerieLangat/Intro-Assignments/master/Copy%20of%20train.csv')
df.head()
# + id="JKv1GOxKn0PV" colab_type="code" colab={}
df.dtypes
# + id="kdUFxWTuws04" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 346} outputId="988cb967-39a5-403d-9351-5241c1661546"
df.describe()
# + id="cHJGoBMJw8Je" colab_type="code" colab={}
df.isna().sum()
# + id="dstt5p4m9Oiw" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 295} outputId="3ff00360-3390-4e31-d1d2-74fd8a66025d"
import matplotlib.pyplot as plt
plt.scatter(df.LotArea, df.YearBuilt)
plt.title('Lot Area and Year Built')
plt.xlabel('Lot Area')
plt.ylabel('Year Built')
plt.show()
# + id="zE6qDuA098O9" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 295} outputId="7afa354e-4621-463e-951f-115fd55836e0"
plt.hist(df['YearBuilt'], bins=20)
plt.xlabel('Year Built')
plt.ylabel('Number of Homes Built')
plt.title("Homes Built")
plt.show()
# + id="WGaI8Kjd-wJe" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 300} outputId="23203763-5c03-4852-d711-fc5346e0d274"
import seaborn as sns
sns.distplot(df.YearBuilt, hist=False)
# + id="bkaJb4_p_Hi0" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 85} outputId="1daf6d57-1077-4fa7-952f-9510daf3f8ca"
graph_3 = sns.pairplot(df)
# + [markdown] id="MZCxTwKuReV9" colab_type="text"
# ## Stretch Goals - Other types and sources of data
#
# Not all data comes in a nice single file - for example, image classification involves handling lots of image files. You still will probably want labels for them, so you may have tabular data in addition to the image blobs - and the images may be reduced in resolution and even fit in a regular csv as a bunch of numbers.
#
# If you're interested in natural language processing and analyzing text, that is another example where, while it can be put in a csv, you may end up loading much larger raw data and generating features that can then be thought of in a more standard tabular fashion.
#
# Overall you will in the course of learning data science deal with loading data in a variety of ways. Another common way to get data is from a database - most modern applications are backed by one or more databases, which you can query to get data to analyze. We'll cover this more in our data engineering unit.
#
# How does data get in the database? Most applications generate logs - text files with lots and lots of records of each use of the application. Databases are often populated based on these files, but in some situations you may directly analyze log files. The usual way to do this is with command line (Unix) tools - command lines are intimidating, so don't expect to learn them all at once, but depending on your interests it can be useful to practice.
#
# One last major source of data is APIs: https://github.com/toddmotto/public-apis
#
# API stands for Application Programming Interface, and while originally meant e.g. the way an application interfaced with the GUI or other aspects of an operating system, now it largely refers to online services that let you query and retrieve data. You can essentially think of most of them as "somebody else's database" - you have (usually limited) access.
#
# *Stretch goal* - research one of the above extended forms of data/data loading. See if you can get a basic example working in a notebook. Image, text, or (public) APIs are probably more tractable - databases are interesting, but there aren't many publicly accessible and they require a great deal of setup.
# + id="f4QP6--JBXNK" colab_type="code" colab={}
| 15,079 |
/phishing.ipynb | 023f1aa4cf7abad7757986940d5d7d6fa5e55f25 | [] | no_license | maciejgorczak/phishing | https://github.com/maciejgorczak/phishing | 0 | 0 | null | null | null | null | Jupyter Notebook | false | false | .py | 42,792 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
import numpy as np
import matplotlib.pyplot as plt
data = np.random.binomial(1, 0.25, (100000, 1000))
f = lambda data, m: np.apply_along_axis(np.mean, 0, data[:m])
means = np.array([f(data[0],m) for m in range(1,1001)])
lay preferences.
# %matplotlib inline
pandas.options.display.float_format = '{:.3f}'.format
# Suppress annoying harmless error.
warnings.filterwarnings(
action="ignore",
module="scipy",
message="^internal gelsd"
)
# -
crimes = pandas.read_csv('table_8.csv', thousands=',')
crimes.fillna(0, inplace=True)
crimes
# # Clean the Data
# +
# Remove empty rows/columns
crimes = crimes[:-3]
crimes = crimes.drop(['Rape1'], axis=1)
data = crimes[['City','Population','Murder', 'Robbery', 'Property_crime']].sort_values(by=['Population'], ascending =False)
# -
# Check for outliers
datag = pandas.melt(data, id_vars=['City'])
datag.head()
graph = seaborn.FacetGrid(datag, col="variable", sharey=False, sharex=False, col_wrap = 5, height = 5, aspect=.5)
graph = (graph.map(seaborn.boxplot, "value"))
graph = graph.set(xscale="log")
# Make a scatterplot matrix.
graph = seaborn.PairGrid(data.dropna(), diag_sharey=False)
# Scatterplot.
graph.map_upper(plt.scatter, alpha=.5)
# Fit line summarizing the linear relationship of the two variables.
graph.map_lower(seaborn.regplot, scatter_kws=dict(alpha=0))
# Give information about the univariate distributions of the variables.
graph.map_diag(seaborn.kdeplot, lw=3)
plt.show()
data.head()
# With New York being so much more dramatically larger than the others in Pop, Pop2, Property_crime trends are difficult/impossible to see.
# +
# Remove outlier (New York)
data1 = data[1:]
data1.head()
# +
# Check data without outlier (New York)
# Check for outliers
data1g = pandas.melt(data1, id_vars=['City'])
data1g.head()
graph = seaborn.FacetGrid(data1g, col="variable", sharey=False, sharex=False, col_wrap = 5, height = 5, aspect=.5)
graph = (graph.map(seaborn.boxplot, "value"))
graph = graph.set(xscale="log")
# +
#714 PairPlot to seaborn.PairGrid
# Make a scatterplot matrix.
graph = seaborn.PairGrid(data1.dropna(), diag_sharey=False)
# Scatterplot.
graph.map_upper(plt.scatter, alpha=.5)
# Fit line summarizing the linear relationship of the two variables.
graph.map_lower(seaborn.regplot, scatter_kws=dict(alpha=0))
# Give information about the univariate distributions of the variables.
graph.map_diag(seaborn.kdeplot, lw=3)
plt.show()
# -
# There are still some significantly larger cities but now other populations are visible, will not remove any further outliers
# ## Generate Features
# +
# Create 2 categorical columns and pop^2
# w/ NYC
data.insert(2,'Pop2', data['Population']**2)
data.insert(3,'Murder_dum',numpy.where(data['Murder']>0, 1, 0))
data.insert(5,'Robbery_dum',numpy.where(data['Robbery']>0, 1, 0))
# w/o NYC
#data1['Pop2'] = data1['Population']**2
data1.insert(2,'Pop2', data1['Population']**2)
data1.insert(3,'Murder_dum',numpy.where(data1['Murder']>0, 1, 0))
data1.insert(6,'Robbery_dum',numpy.where(data1['Robbery']>0, 1, 0))
#data = crimes[['Population','Pop2','Murder_dum', 'Robbery_dum', 'Property_crime']].sort_values(by=['Population'], ascending =False)
# -
data1
# ## Generate Model
# +
# function for linear reg modeling
def lin_model(rdata, target):
# Instantiate and fit our model.
regr = linear_model.LinearRegression()
regr.fit(rdata, target)
# Inspect the results.
print('\nCoefficients: \n', regr.coef_)
print('\nIntercept: \n', regr.intercept_)
print('\nR-squared: \n', regr.score(rdata, target))
predicted = regr.predict(rdata).ravel()
actual = target.ravel()
# Calculate the error
residual = actual - predicted
# Error historgram
plt.hist(residual)
plt.title('Residual Counts')
plt.xlabel('Residual')
plt.ylabel('Count')
plt.show()
# Error scatter plot
plt.scatter(predicted, residual)
plt.xlabel('Predicted')
plt.ylabel('Residual')
plt.axhline(y=0)
plt.title('Residual vs. Predicted')
plt.show()
# +
target = data['Property_crime'].values.reshape(-1, 1)
rdata = data[['Population', 'Pop2', 'Murder_dum', 'Robbery_dum']]
lin_model(rdata, target)
# +
target = data1['Property_crime'].values.reshape(-1, 1)
rdata = data1[['Population', 'Pop2', 'Murder_dum', 'Robbery_dum']]
lin_model(rdata, target)
c4ac0-4229-4e56-d9e1-6d19d311ea56" colab={"base_uri": "https://localhost:8080/", "height": 34}
#splitting data into training and test datasets
mask = np.random.rand(len(data)) < 0.75
train = data[mask]
test = data[~mask]
print(train.shape, test.shape)
# + id="LvawbvcqVUVJ" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 1000} outputId="eba90100-f3ea-4b29-fc7c-add7e2e0764a"
'''
almost manual tuning of neural network
'''
hiperparameters = [32, 64, 128]
batch = [30, 50, 70]
learning = [0.007, 0.009, 0.01]
di = {}
for i in hiperparameters:
'''
hiperparameters of the neural network
'''
inputSize = len(train.columns) -1
hidden1Size = i
hidden2Size = i
numClasses = 2
numEpoch = 100
print(i)
bat = {}
for b in batch:
batchSize = b
print(b)
ler = {}
for l in learning:
learningRate = l
print(l)
trainLoader = torch.utils.data.DataLoader(dataset=torch.tensor(train.values), batch_size=batchSize, shuffle=True)
testLoader = torch.utils.data.DataLoader(dataset=torch.tensor(test.values))
'''
below model of the neural network
'''
class DeepNeuralNetwork(nn.Module):
def __init__(self, inputSize, hidden1Size, hidden2Size, numClasses):
super(DeepNeuralNetwork, self).__init__()
self.fc1 = nn.Linear(inputSize, hidden1Size)
self.relu1 = nn.ReLU()
self.fc2 = nn.Linear(hidden1Size, hidden2Size)
self.relu2 = nn.ReLU()
self.fc3 = nn.Linear(hidden2Size, numClasses)
self.logsm1 = nn.LogSoftmax(dim=1)
def forward(self, x):
out = self.fc1(x)
out = self.relu1(out)
out = self.fc2(out)
out = self.relu2(out)
out = self.fc3(out)
out = self.logsm1(out)
return out
dnn = DeepNeuralNetwork(inputSize, hidden1Size, hidden2Size, numClasses)
#loss function and optimiizer
lossFN = nn.NLLLoss()
optimizer = torch.optim.Adam(dnn.parameters(), lr=learningRate)
#training
for epoch in range(0, numEpoch):
for i, data in enumerate(trainLoader,0):
labels = Variable(data[:,-1])
data = Variable(data[:,0:22].float())
optimizer.zero_grad()
outputs = dnn(data)
loss = lossFN(outputs, labels.long())
loss.backward()
optimizer.step()
#print('Epoch [{}/{}], Loss: {:.4f}'.format(epoch+1, numEpoch, loss.item()))
#checking the model on test data
correct = 0
total = 0
for data in testLoader:
labels = Variable(data[:,-1])
data = Variable(data[:,0:22].float())
outputs = dnn(data)
_, predicted = torch.max(outputs.data, 1)
total += labels.size(0)
correct += int((predicted == labels.long()).sum())
print('Accuracy of the network on the data: {:.4f}%'.format(100 * correct / total))
bas = float("{:.4f}".format(100 * correct / total))
ler[l] = bas
bat[b] = ler
di[i] = bat
# + id="MoawwQFtY_vX" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 34} outputId="d1fe62d3-49a7-450d-ba87-681ca214ce43"
print(di)
_iter2 = KMeans(n_clusters=5, init="random", n_init=1,
algorithm="full", max_iter=2, random_state=1)
kmeans_iter3 = KMeans(n_clusters=5, init="random", n_init=1,
algorithm="full", max_iter=3, random_state=1)
kmeans_iter1.fit(X)
kmeans_iter2.fit(X)
kmeans_iter3.fit(X)
# And let's plot this:
# +
plt.figure(figsize=(10, 8))
plt.subplot(321)
plot_data(X)
plot_centroids(kmeans_iter1.cluster_centers_, circle_color='r', cross_color='w')
plt.ylabel("$x_2$", fontsize=14, rotation=0)
plt.tick_params(labelbottom=False)
plt.title("Update the centroids (initially randomly)", fontsize=14)
plt.subplot(322)
plot_decision_boundaries(kmeans_iter1, X, show_xlabels=False, show_ylabels=False)
plt.title("Label the instances", fontsize=14)
plt.subplot(323)
plot_decision_boundaries(kmeans_iter1, X, show_centroids=False, show_xlabels=False)
plot_centroids(kmeans_iter2.cluster_centers_)
plt.subplot(324)
plot_decision_boundaries(kmeans_iter2, X, show_xlabels=False, show_ylabels=False)
plt.subplot(325)
plot_decision_boundaries(kmeans_iter2, X, show_centroids=False)
plot_centroids(kmeans_iter3.cluster_centers_)
plt.subplot(326)
plot_decision_boundaries(kmeans_iter3, X, show_ylabels=False)
save_fig("kmeans_algorithm_plot")
plt.show()
# -
# ### K-Means Variability
# In the original K-Means algorithm, the centroids are just initialized randomly, and the algorithm simply runs a single iteration to gradually improve the centroids, as we saw above.
#
# However, one major problem with this approach is that if you run K-Means multiple times (or with different random seeds), it can converge to very different solutions, as you can see below:
def plot_clusterer_comparison(clusterer1, clusterer2, X, title1=None, title2=None):
clusterer1.fit(X)
clusterer2.fit(X)
plt.figure(figsize=(10, 3.2))
plt.subplot(121)
plot_decision_boundaries(clusterer1, X)
if title1:
plt.title(title1, fontsize=14)
plt.subplot(122)
plot_decision_boundaries(clusterer2, X, show_ylabels=False)
if title2:
plt.title(title2, fontsize=14)
# +
kmeans_rnd_init1 = KMeans(n_clusters=5, init="random", n_init=1,
algorithm="full", random_state=11)
kmeans_rnd_init2 = KMeans(n_clusters=5, init="random", n_init=1,
algorithm="full", random_state=19)
plot_clusterer_comparison(kmeans_rnd_init1, kmeans_rnd_init2, X,
"Solution 1", "Solution 2 (with a different random init)")
save_fig("kmeans_variability_plot")
plt.show()
# -
# ### Inertia
# To select the best model, we will need a way to evaluate a K-Mean model's performance. Unfortunately, clustering is an unsupervised task, so we do not have the targets. But at least we can measure the distance between each instance and its centroid. This is the idea behind the _inertia_ metric:
kmeans.inertia_
# As you can easily verify, inertia is the sum of the squared distances between each training instance and its closest centroid:
X_dist = kmeans.transform(X)
np.sum(X_dist[np.arange(len(X_dist)), kmeans.labels_]**2)
# The `score()` method returns the negative inertia. Why negative? Well, it is because a predictor's `score()` method must always respect the "_great is better_" rule.
kmeans.score(X)
# ### Multiple Initializations
# So one approach to solve the variability issue is to simply run the K-Means algorithm multiple times with different random initializations, and select the solution that minimizes the inertia. For example, here are the inertias of the two "bad" models shown in the previous figure:
kmeans_rnd_init1.inertia_
kmeans_rnd_init2.inertia_
# As you can see, they have a higher inertia than the first "good" model we trained, which means they are probably worse.
# When you set the `n_init` hyperparameter, Scikit-Learn runs the original algorithm `n_init` times, and selects the solution that minimizes the inertia. By default, Scikit-Learn sets `n_init=10`.
kmeans_rnd_10_inits = KMeans(n_clusters=5, init="random", n_init=10,
algorithm="full", random_state=11)
kmeans_rnd_10_inits.fit(X)
# As you can see, we end up with the initial model, which is certainly the optimal K-Means solution (at least in terms of inertia, and assuming $k=5$).
plt.figure(figsize=(8, 4))
plot_decision_boundaries(kmeans_rnd_10_inits, X)
plt.show()
# ### K-Means++
# Instead of initializing the centroids entirely randomly, it is preferable to initialize them using the following algorithm, proposed in a [2006 paper](https://goo.gl/eNUPw6) by David Arthur and Sergei Vassilvitskii:
# * Take one centroid $c_1$, chosen uniformly at random from the dataset.
# * Take a new center $c_i$, choosing an instance $\mathbf{x}_i$ with probability: $D(\mathbf{x}_i)^2$ / $\sum\limits_{j=1}^{m}{D(\mathbf{x}_j)}^2$ where $D(\mathbf{x}_i)$ is the distance between the instance $\mathbf{x}_i$ and the closest centroid that was already chosen. This probability distribution ensures that instances that are further away from already chosen centroids are much more likely be selected as centroids.
# * Repeat the previous step until all $k$ centroids have been chosen.
# The rest of the K-Means++ algorithm is just regular K-Means. With this initialization, the K-Means algorithm is much less likely to converge to a suboptimal solution, so it is possible to reduce `n_init` considerably. Most of the time, this largely compensates for the additional complexity of the initialization process.
# To set the initialization to K-Means++, simply set `init="k-means++"` (this is actually the default):
KMeans()
good_init = np.array([[-3, 3], [-3, 2], [-3, 1], [-1, 2], [0, 2]])
kmeans = KMeans(n_clusters=5, init=good_init, n_init=1, random_state=42)
kmeans.fit(X)
kmeans.inertia_
# ### Accelerated K-Means
# The K-Means algorithm can be significantly accelerated by avoiding many unnecessary distance calculations: this is achieved by exploiting the triangle inequality (given three points A, B and C, the distance AC is always such that AC ≤ AB + BC) and by keeping track of lower and upper bounds for distances between instances and centroids (see this [2003 paper](https://www.aaai.org/Papers/ICML/2003/ICML03-022.pdf) by Charles Elkan for more details).
# To use Elkan's variant of K-Means, just set `algorithm="elkan"`. Note that it does not support sparse data, so by default, Scikit-Learn uses `"elkan"` for dense data, and `"full"` (the regular K-Means algorithm) for sparse data.
# %timeit -n 50 KMeans(algorithm="elkan").fit(X)
# %timeit -n 50 KMeans(algorithm="full").fit(X)
# ### Mini-Batch K-Means
# Scikit-Learn also implements a variant of the K-Means algorithm that supports mini-batches (see [this paper](http://www.eecs.tufts.edu/~dsculley/papers/fastkmeans.pdf)):
from sklearn.cluster import MiniBatchKMeans
minibatch_kmeans = MiniBatchKMeans(n_clusters=5, random_state=42)
minibatch_kmeans.fit(X)
minibatch_kmeans.inertia_
# If the dataset does not fit in memory, the simplest option is to use the `memmap` class, just like we did for incremental PCA in the previous chapter. First let's load MNIST:
# +
import urllib
from sklearn.datasets import fetch_openml
mnist = fetch_openml('mnist_784', version=1)
mnist.target = mnist.target.astype(np.int64)
# +
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(
mnist["data"], mnist["target"], random_state=42)
# -
# Next, let's write it to a `memmap`:
filename = "my_mnist.data"
X_mm = np.memmap(filename, dtype='float32', mode='write', shape=X_train.shape)
X_mm[:] = X_train
minibatch_kmeans = MiniBatchKMeans(n_clusters=10, batch_size=10, random_state=42)
minibatch_kmeans.fit(X_mm)
# If your data is so large that you cannot use `memmap`, things get more complicated. Let's start by writing a function to load the next batch (in real life, you would load the data from disk):
def load_next_batch(batch_size):
return X[np.random.choice(len(X), batch_size, replace=False)]
# Now we can train the model by feeding it one batch at a time. We also need to implement multiple initializations and keep the model with the lowest inertia:
np.random.seed(42)
# +
k = 5
n_init = 10
n_iterations = 100
batch_size = 100
init_size = 500 # more data for K-Means++ initialization
evaluate_on_last_n_iters = 10
best_kmeans = None
for init in range(n_init):
minibatch_kmeans = MiniBatchKMeans(n_clusters=k, init_size=init_size)
X_init = load_next_batch(init_size)
minibatch_kmeans.partial_fit(X_init)
minibatch_kmeans.sum_inertia_ = 0
for iteration in range(n_iterations):
X_batch = load_next_batch(batch_size)
minibatch_kmeans.partial_fit(X_batch)
if iteration >= n_iterations - evaluate_on_last_n_iters:
minibatch_kmeans.sum_inertia_ += minibatch_kmeans.inertia_
if (best_kmeans is None or
minibatch_kmeans.sum_inertia_ < best_kmeans.sum_inertia_):
best_kmeans = minibatch_kmeans
# -
best_kmeans.score(X)
# Mini-batch K-Means is much faster than regular K-Means:
# %timeit KMeans(n_clusters=5).fit(X)
# %timeit MiniBatchKMeans(n_clusters=5).fit(X)
# That's *much* faster! However, its performance is often lower (higher inertia), and it keeps degrading as _k_ increases. Let's plot the inertia ratio and the training time ratio between Mini-batch K-Means and regular K-Means:
from timeit import timeit
times = np.empty((100, 2))
inertias = np.empty((100, 2))
for k in range(1, 101):
kmeans_ = KMeans(n_clusters=k, random_state=42)
minibatch_kmeans = MiniBatchKMeans(n_clusters=k, random_state=42)
print("\r{}/{}".format(k, 100), end="")
times[k-1, 0] = timeit("kmeans_.fit(X)", number=10, globals=globals())
times[k-1, 1] = timeit("minibatch_kmeans.fit(X)", number=10, globals=globals())
inertias[k-1, 0] = kmeans_.inertia_
inertias[k-1, 1] = minibatch_kmeans.inertia_
# +
plt.figure(figsize=(10,4))
plt.subplot(121)
plt.plot(range(1, 101), inertias[:, 0], "r--", label="K-Means")
plt.plot(range(1, 101), inertias[:, 1], "b.-", label="Mini-batch K-Means")
plt.xlabel("$k$", fontsize=16)
plt.title("Inertia", fontsize=14)
plt.legend(fontsize=14)
plt.axis([1, 100, 0, 100])
plt.subplot(122)
plt.plot(range(1, 101), times[:, 0], "r--", label="K-Means")
plt.plot(range(1, 101), times[:, 1], "b.-", label="Mini-batch K-Means")
plt.xlabel("$k$", fontsize=16)
plt.title("Training time (seconds)", fontsize=14)
plt.axis([1, 100, 0, 6])
save_fig("minibatch_kmeans_vs_kmeans")
plt.show()
# -
# ### Finding the optimal number of clusters
# What if the number of clusters was set to a lower or greater value than 5?
# +
kmeans_k3 = KMeans(n_clusters=3, random_state=42)
kmeans_k8 = KMeans(n_clusters=8, random_state=42)
plot_clusterer_comparison(kmeans_k3, kmeans_k8, X, "$k=3$", "$k=8$")
save_fig("bad_n_clusters_plot")
plt.show()
# -
# Ouch, these two models don't look great. What about their inertias?
kmeans_k3.inertia_
kmeans_k8.inertia_
# No, we cannot simply take the value of $k$ that minimizes the inertia, since it keeps getting lower as we increase $k$. Indeed, the more clusters there are, the closer each instance will be to its closest centroid, and therefore the lower the inertia will be. However, we can plot the inertia as a function of $k$ and analyze the resulting curve:
kmeans_per_k = [KMeans(n_clusters=k, random_state=42).fit(X)
for k in range(1, 10)]
inertias = [model.inertia_ for model in kmeans_per_k]
plt.figure(figsize=(8, 3.5))
plt.plot(range(1, 10), inertias, "bo-")
plt.xlabel("$k$", fontsize=14)
plt.ylabel("Inertia", fontsize=14)
plt.annotate('Elbow',
xy=(4, inertias[3]),
xytext=(0.55, 0.55),
textcoords='figure fraction',
fontsize=16,
arrowprops=dict(facecolor='black', shrink=0.1)
)
plt.axis([1, 8.5, 0, 1300])
save_fig("inertia_vs_k_plot")
plt.show()
# As you can see, there is an elbow at $k=4$, which means that less clusters than that would be bad, and more clusters would not help much and might cut clusters in half. So $k=4$ is a pretty good choice. Of course in this example it is not perfect since it means that the two blobs in the lower left will be considered as just a single cluster, but it's a pretty good clustering nonetheless.
plot_decision_boundaries(kmeans_per_k[4-1], X)
plt.show()
# Another approach is to look at the _silhouette score_, which is the mean _silhouette coefficient_ over all the instances. An instance's silhouette coefficient is equal to $(b - a)/\max(a, b)$ where $a$ is the mean distance to the other instances in the same cluster (it is the _mean intra-cluster distance_), and $b$ is the _mean nearest-cluster distance_, that is the mean distance to the instances of the next closest cluster (defined as the one that minimizes $b$, excluding the instance's own cluster). The silhouette coefficient can vary between -1 and +1: a coefficient close to +1 means that the instance is well inside its own cluster and far from other clusters, while a coefficient close to 0 means that it is close to a cluster boundary, and finally a coefficient close to -1 means that the instance may have been assigned to the wrong cluster.
# Let's plot the silhouette score as a function of $k$:
from sklearn.metrics import silhouette_score
silhouette_score(X, kmeans.labels_)
silhouette_scores = [silhouette_score(X, model.labels_)
for model in kmeans_per_k[1:]]
plt.figure(figsize=(8, 3))
plt.plot(range(2, 10), silhouette_scores, "bo-")
plt.xlabel("$k$", fontsize=14)
plt.ylabel("Silhouette score", fontsize=14)
plt.axis([1.8, 8.5, 0.55, 0.7])
save_fig("silhouette_score_vs_k_plot")
plt.show()
# As you can see, this visualization is much richer than the previous one: in particular, although it confirms that $k=4$ is a very good choice, but it also underlines the fact that $k=5$ is quite good as well.
# An even more informative visualization is given when you plot every instance's silhouette coefficient, sorted by the cluster they are assigned to and by the value of the coefficient. This is called a _silhouette diagram_:
# +
from sklearn.metrics import silhouette_samples
from matplotlib.ticker import FixedLocator, FixedFormatter
plt.figure(figsize=(11, 9))
for k in (3, 4, 5, 6):
plt.subplot(2, 2, k - 2)
y_pred = kmeans_per_k[k - 1].labels_
silhouette_coefficients = silhouette_samples(X, y_pred)
padding = len(X) // 30
pos = padding
ticks = []
for i in range(k):
coeffs = silhouette_coefficients[y_pred == i]
coeffs.sort()
color = mpl.cm.Spectral(i / k)
plt.fill_betweenx(np.arange(pos, pos + len(coeffs)), 0, coeffs,
facecolor=color, edgecolor=color, alpha=0.7)
ticks.append(pos + len(coeffs) // 2)
pos += len(coeffs) + padding
plt.gca().yaxis.set_major_locator(FixedLocator(ticks))
plt.gca().yaxis.set_major_formatter(FixedFormatter(range(k)))
if k in (3, 5):
plt.ylabel("Cluster")
if k in (5, 6):
plt.gca().set_xticks([-0.1, 0, 0.2, 0.4, 0.6, 0.8, 1])
plt.xlabel("Silhouette Coefficient")
else:
plt.tick_params(labelbottom=False)
plt.axvline(x=silhouette_scores[k - 2], color="red", linestyle="--")
plt.title("$k={}$".format(k), fontsize=16)
save_fig("silhouette_analysis_plot")
plt.show()
# -
# ### Limits of K-Means
X1, y1 = make_blobs(n_samples=1000, centers=((4, -4), (0, 0)), random_state=42)
X1 = X1.dot(np.array([[0.374, 0.95], [0.732, 0.598]]))
X2, y2 = make_blobs(n_samples=250, centers=1, random_state=42)
X2 = X2 + [6, -8]
X = np.r_[X1, X2]
y = np.r_[y1, y2]
plot_clusters(X)
kmeans_good = KMeans(n_clusters=3, init=np.array([[-1.5, 2.5], [0.5, 0], [4, 0]]), n_init=1, random_state=42)
kmeans_bad = KMeans(n_clusters=3, random_state=42)
kmeans_good.fit(X)
kmeans_bad.fit(X)
# +
plt.figure(figsize=(10, 3.2))
plt.subplot(121)
plot_decision_boundaries(kmeans_good, X)
plt.title("Inertia = {:.1f}".format(kmeans_good.inertia_), fontsize=14)
plt.subplot(122)
plot_decision_boundaries(kmeans_bad, X, show_ylabels=False)
plt.title("Inertia = {:.1f}".format(kmeans_bad.inertia_), fontsize=14)
save_fig("bad_kmeans_plot")
plt.show()
# -
# ### Using clustering for image segmentation
# Download the ladybug image
images_path = os.path.join(PROJECT_ROOT_DIR, "images", "unsupervised_learning")
os.makedirs(images_path, exist_ok=True)
DOWNLOAD_ROOT = "https://raw.githubusercontent.com/ageron/handson-ml2/master/"
filename = "ladybug.png"
print("Downloading", filename)
url = DOWNLOAD_ROOT + "images/unsupervised_learning/" + filename
urllib.request.urlretrieve(url, os.path.join(images_path, filename))
from matplotlib.image import imread
image = imread(os.path.join(images_path, filename))
image.shape
X = image.reshape(-1, 3)
kmeans = KMeans(n_clusters=8, random_state=42).fit(X)
segmented_img = kmeans.cluster_centers_[kmeans.labels_]
segmented_img = segmented_img.reshape(image.shape)
segmented_imgs = []
n_colors = (10, 8, 6, 4, 2)
for n_clusters in n_colors:
kmeans = KMeans(n_clusters=n_clusters, random_state=42).fit(X)
segmented_img = kmeans.cluster_centers_[kmeans.labels_]
segmented_imgs.append(segmented_img.reshape(image.shape))
# +
plt.figure(figsize=(10,5))
plt.subplots_adjust(wspace=0.05, hspace=0.1)
plt.subplot(231)
plt.imshow(image)
plt.title("Original image")
plt.axis('off')
for idx, n_clusters in enumerate(n_colors):
plt.subplot(232 + idx)
plt.imshow(segmented_imgs[idx])
plt.title("{} colors".format(n_clusters))
plt.axis('off')
save_fig('image_segmentation_diagram', tight_layout=False)
plt.show()
# -
# ### Using Clustering for Preprocessing
# Let's tackle the _digits dataset_ which is a simple MNIST-like dataset containing 1,797 grayscale 8×8 images representing digits 0 to 9.
from sklearn.datasets import load_digits
X_digits, y_digits = load_digits(return_X_y=True)
# Let's split it into a training set and a test set:
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X_digits, y_digits, random_state=42)
# Now let's fit a Logistic Regression model and evaluate it on the test set:
from sklearn.linear_model import LogisticRegression
log_reg = LogisticRegression(multi_class="ovr", solver="lbfgs", max_iter=5000, random_state=42)
log_reg.fit(X_train, y_train)
log_reg.score(X_test, y_test)
# Okay, that's our baseline: 96.89% accuracy. Let's see if we can do better by using K-Means as a preprocessing step. We will create a pipeline that will first cluster the training set into 50 clusters and replace the images with their distances to the 50 clusters, then apply a logistic regression model:
from sklearn.pipeline import Pipeline
pipeline = Pipeline([
("kmeans", KMeans(n_clusters=50, random_state=42)),
("log_reg", LogisticRegression(multi_class="ovr", solver="lbfgs", max_iter=5000, random_state=42)),
])
pipeline.fit(X_train, y_train)
pipeline.score(X_test, y_test)
1 - (1 - 0.977777) / (1 - 0.968888)
# How about that? We reduced the error rate by over 28%! But we chose the number of clusters $k$ completely arbitrarily, we can surely do better. Since K-Means is just a preprocessing step in a classification pipeline, finding a good value for $k$ is much simpler than earlier: there's no need to perform silhouette analysis or minimize the inertia, the best value of $k$ is simply the one that results in the best classification performance.
from sklearn.model_selection import GridSearchCV
param_grid = dict(kmeans__n_clusters=range(2, 100))
grid_clf = GridSearchCV(pipeline, param_grid, cv=3, verbose=2)
grid_clf.fit(X_train, y_train)
# Let's see what the best number of clusters is:
grid_clf.best_params_
grid_clf.score(X_test, y_test)
# ### Clustering for Semi-supervised Learning
# Another use case for clustering is in semi-supervised learning, when we have plenty of unlabeled instances and very few labeled instances.
# Let's look at the performance of a logistic regression model when we only have 50 labeled instances:
n_labeled = 50
log_reg = LogisticRegression(multi_class="ovr", solver="lbfgs", random_state=42)
log_reg.fit(X_train[:n_labeled], y_train[:n_labeled])
log_reg.score(X_test, y_test)
# It's much less than earlier of course. Let's see how we can do better. First, let's cluster the training set into 50 clusters, then for each cluster let's find the image closest to the centroid. We will call these images the representative images:
k = 50
kmeans = KMeans(n_clusters=k, random_state=42)
X_digits_dist = kmeans.fit_transform(X_train)
representative_digit_idx = np.argmin(X_digits_dist, axis=0)
X_representative_digits = X_train[representative_digit_idx]
# Now let's plot these representative images and label them manually:
# +
plt.figure(figsize=(8, 2))
for index, X_representative_digit in enumerate(X_representative_digits):
plt.subplot(k // 10, 10, index + 1)
plt.imshow(X_representative_digit.reshape(8, 8), cmap="binary", interpolation="bilinear")
plt.axis('off')
save_fig("representative_images_diagram", tight_layout=False)
plt.show()
# -
y_representative_digits = np.array([
4, 8, 0, 6, 8, 3, 7, 7, 9, 2,
5, 5, 8, 5, 2, 1, 2, 9, 6, 1,
1, 6, 9, 0, 8, 3, 0, 7, 4, 1,
6, 5, 2, 4, 1, 8, 6, 3, 9, 2,
4, 2, 9, 4, 7, 6, 2, 3, 1, 1])
# Now we have a dataset with just 50 labeled instances, but instead of being completely random instances, each of them is a representative image of its cluster. Let's see if the performance is any better:
log_reg = LogisticRegression(multi_class="ovr", solver="lbfgs", max_iter=5000, random_state=42)
log_reg.fit(X_representative_digits, y_representative_digits)
log_reg.score(X_test, y_test)
# Wow! We jumped from 83.3% accuracy to 92.2%, although we are still only training the model on 50 instances. Since it's often costly and painful to label instances, especially when it has to be done manually by experts, it's a good idea to make them label representative instances rather than just random instances.
# But perhaps we can go one step further: what if we propagated the labels to all the other instances in the same cluster?
y_train_propagated = np.empty(len(X_train), dtype=np.int32)
for i in range(k):
y_train_propagated[kmeans.labels_==i] = y_representative_digits[i]
log_reg = LogisticRegression(multi_class="ovr", solver="lbfgs", max_iter=5000, random_state=42)
log_reg.fit(X_train, y_train_propagated)
log_reg.score(X_test, y_test)
# We got a tiny little accuracy boost. Better than nothing, but we should probably have propagated the labels only to the instances closest to the centroid, because by propagating to the full cluster, we have certainly included some outliers. Let's only propagate the labels to the 20th percentile closest to the centroid:
# +
percentile_closest = 20
X_cluster_dist = X_digits_dist[np.arange(len(X_train)), kmeans.labels_]
for i in range(k):
in_cluster = (kmeans.labels_ == i)
cluster_dist = X_cluster_dist[in_cluster]
cutoff_distance = np.percentile(cluster_dist, percentile_closest)
above_cutoff = (X_cluster_dist > cutoff_distance)
X_cluster_dist[in_cluster & above_cutoff] = -1
# -
partially_propagated = (X_cluster_dist != -1)
X_train_partially_propagated = X_train[partially_propagated]
y_train_partially_propagated = y_train_propagated[partially_propagated]
log_reg = LogisticRegression(multi_class="ovr", solver="lbfgs", max_iter=5000, random_state=42)
log_reg.fit(X_train_partially_propagated, y_train_partially_propagated)
log_reg.score(X_test, y_test)
# Nice! With just 50 labeled instances (just 5 examples per class on average!), we got 94% performance, which is pretty close to the performance of logistic regression on the fully labeled _digits_ dataset (which was 96.9%).
# This is because the propagated labels are actually pretty good: their accuracy is very close to 99%:
np.mean(y_train_partially_propagated == y_train[partially_propagated])
# You could now do a few iterations of *active learning*:
# 1. Manually label the instances that the classifier is least sure about, if possible by picking them in distinct clusters.
# 2. Train a new model with these additional labels.
# ## DBSCAN
from sklearn.datasets import make_moons
X, y = make_moons(n_samples=1000, noise=0.05, random_state=42)
X
y
from sklearn.cluster import DBSCAN
dbscan = DBSCAN(eps=0.05, min_samples=5)
dbscan.fit(X)
dbscan.labels_[:10]
len(dbscan.core_sample_indices_)
dbscan.core_sample_indices_[:10]
dbscan.components_[:3]
np.unique(dbscan.labels_)
dbscan2 = DBSCAN(eps=0.2)
dbscan2.fit(X)
def plot_dbscan(dbscan, X, size, show_xlabels=True, show_ylabels=True):
core_mask = np.zeros_like(dbscan.labels_, dtype=bool)
core_mask[dbscan.core_sample_indices_] = True
anomalies_mask = dbscan.labels_ == -1
non_core_mask = ~(core_mask | anomalies_mask)
cores = dbscan.components_
anomalies = X[anomalies_mask]
non_cores = X[non_core_mask]
plt.scatter(cores[:, 0], cores[:, 1],
c=dbscan.labels_[core_mask], marker='o', s=size, cmap="Paired")
plt.scatter(cores[:, 0], cores[:, 1], marker='*', s=20, c=dbscan.labels_[core_mask])
plt.scatter(anomalies[:, 0], anomalies[:, 1],
c="r", marker="x", s=100)
plt.scatter(non_cores[:, 0], non_cores[:, 1], c=dbscan.labels_[non_core_mask], marker=".")
if show_xlabels:
plt.xlabel("$x_1$", fontsize=14)
else:
plt.tick_params(labelbottom=False)
if show_ylabels:
plt.ylabel("$x_2$", fontsize=14, rotation=0)
else:
plt.tick_params(labelleft=False)
plt.title("eps={:.2f}, min_samples={}".format(dbscan.eps, dbscan.min_samples), fontsize=14)
# +
plt.figure(figsize=(9, 3.2))
plt.subplot(121)
plot_dbscan(dbscan, X, size=100)
plt.subplot(122)
plot_dbscan(dbscan2, X, size=600, show_ylabels=False)
save_fig("dbscan_plot")
plt.show()
# -
dbscan = dbscan2
from sklearn.neighbors import KNeighborsClassifier
knn = KNeighborsClassifier(n_neighbors=50)
knn.fit(dbscan.components_, dbscan.labels_[dbscan.core_sample_indices_])
X_new = np.array([[-0.5, 0], [0, 0.5], [1, -0.1], [2, 1]])
knn.predict(X_new)
knn.predict_proba(X_new)
plt.figure(figsize=(6, 3))
plot_decision_boundaries(knn, X, show_centroids=False)
plt.scatter(X_new[:, 0], X_new[:, 1], c="b", marker="+", s=200, zorder=10)
save_fig("cluster_classification_plot")
plt.show()
y_dist, y_pred_idx = knn.kneighbors(X_new, n_neighbors=1)
y_pred = dbscan.labels_[dbscan.core_sample_indices_][y_pred_idx]
y_pred[y_dist > 0.2] = -1
y_pred.ravel()
# ## Other Clustering Algorithms
# ### Spectral Clustering
from sklearn.cluster import SpectralClustering
sc1 = SpectralClustering(n_clusters=2, gamma=100, random_state=42)
sc1.fit(X)
sc2 = SpectralClustering(n_clusters=2, gamma=1, random_state=42)
sc2.fit(X)
np.percentile(sc1.affinity_matrix_, 95)
def plot_spectral_clustering(sc, X, size, alpha, show_xlabels=True, show_ylabels=True):
plt.scatter(X[:, 0], X[:, 1], marker='o', s=size, c='gray', cmap="Paired", alpha=alpha)
plt.scatter(X[:, 0], X[:, 1], marker='o', s=30, c='w')
plt.scatter(X[:, 0], X[:, 1], marker='.', s=10, c=sc.labels_, cmap="Paired")
if show_xlabels:
plt.xlabel("$x_1$", fontsize=14)
else:
plt.tick_params(labelbottom=False)
if show_ylabels:
plt.ylabel("$x_2$", fontsize=14, rotation=0)
else:
plt.tick_params(labelleft=False)
plt.title("RBF gamma={}".format(sc.gamma), fontsize=14)
# +
plt.figure(figsize=(9, 3.2))
plt.subplot(121)
plot_spectral_clustering(sc1, X, size=500, alpha=0.1)
plt.subplot(122)
plot_spectral_clustering(sc2, X, size=4000, alpha=0.01, show_ylabels=False)
plt.show()
# -
# ### Agglomerative Clustering
from sklearn.cluster import AgglomerativeClustering
X = np.array([0, 2, 5, 8.5]).reshape(-1, 1)
agg = AgglomerativeClustering(linkage="complete").fit(X)
def learned_parameters(estimator):
return [attrib for attrib in dir(estimator)
if attrib.endswith("_") and not attrib.startswith("_")]
learned_parameters(agg)
agg.children_
# # Gaussian Mixtures
X1, y1 = make_blobs(n_samples=1000, centers=((4, -4), (0, 0)), random_state=42)
X1 = X1.dot(np.array([[0.374, 0.95], [0.732, 0.598]]))
X2, y2 = make_blobs(n_samples=250, centers=1, random_state=42)
X2 = X2 + [6, -8]
X = np.r_[X1, X2]
y = np.r_[y1, y2]
# Let's train a Gaussian mixture model on the previous dataset:
from sklearn.mixture import GaussianMixture
gm = GaussianMixture(n_components=3, n_init=10, random_state=42)
gm.fit(X)
# Let's look at the parameters that the EM algorithm estimated:
gm.weights_
gm.means_
gm.covariances_
# Did the algorithm actually converge?
gm.converged_
# Yes, good. How many iterations did it take?
gm.n_iter_
# You can now use the model to predict which cluster each instance belongs to (hard clustering) or the probabilities that it came from each cluster. For this, just use `predict()` method or the `predict_proba()` method:
gm.predict(X)
gm.predict_proba(X)
# This is a generative model, so you can sample new instances from it (and get their labels):
X_new, y_new = gm.sample(6)
X_new
y_new
# Notice that they are sampled sequentially from each cluster.
# You can also estimate the log of the _probability density function_ (PDF) at any location using the `score_samples()` method:
gm.score_samples(X)
# Let's check that the PDF integrates to 1 over the whole space. We just take a large square around the clusters, and chop it into a grid of tiny squares, then we compute the approximate probability that the instances will be generated in each tiny square (by multiplying the PDF at one corner of the tiny square by the area of the square), and finally summing all these probabilities). The result is very close to 1:
# +
resolution = 100
grid = np.arange(-10, 10, 1 / resolution)
xx, yy = np.meshgrid(grid, grid)
X_full = np.vstack([xx.ravel(), yy.ravel()]).T
pdf = np.exp(gm.score_samples(X_full))
pdf_probas = pdf * (1 / resolution) ** 2
pdf_probas.sum()
# -
# Now let's plot the resulting decision boundaries (dashed lines) and density contours:
# +
from matplotlib.colors import LogNorm
def plot_gaussian_mixture(clusterer, X, resolution=1000, show_ylabels=True):
mins = X.min(axis=0) - 0.1
maxs = X.max(axis=0) + 0.1
xx, yy = np.meshgrid(np.linspace(mins[0], maxs[0], resolution),
np.linspace(mins[1], maxs[1], resolution))
Z = -clusterer.score_samples(np.c_[xx.ravel(), yy.ravel()])
Z = Z.reshape(xx.shape)
plt.contourf(xx, yy, Z,
norm=LogNorm(vmin=1.0, vmax=30.0),
levels=np.logspace(0, 2, 12))
plt.contour(xx, yy, Z,
norm=LogNorm(vmin=1.0, vmax=30.0),
levels=np.logspace(0, 2, 12),
linewidths=1, colors='k')
Z = clusterer.predict(np.c_[xx.ravel(), yy.ravel()])
Z = Z.reshape(xx.shape)
plt.contour(xx, yy, Z,
linewidths=2, colors='r', linestyles='dashed')
plt.plot(X[:, 0], X[:, 1], 'k.', markersize=2)
plot_centroids(clusterer.means_, clusterer.weights_)
plt.xlabel("$x_1$", fontsize=14)
if show_ylabels:
plt.ylabel("$x_2$", fontsize=14, rotation=0)
else:
plt.tick_params(labelleft=False)
# +
plt.figure(figsize=(8, 4))
plot_gaussian_mixture(gm, X)
save_fig("gaussian_mixtures_plot")
plt.show()
# -
# You can impose constraints on the covariance matrices that the algorithm looks for by setting the `covariance_type` hyperparameter:
# * `"full"` (default): no constraint, all clusters can take on any ellipsoidal shape of any size.
# * `"tied"`: all clusters must have the same shape, which can be any ellipsoid (i.e., they all share the same covariance matrix).
# * `"spherical"`: all clusters must be spherical, but they can have different diameters (i.e., different variances).
# * `"diag"`: clusters can take on any ellipsoidal shape of any size, but the ellipsoid's axes must be parallel to the axes (i.e., the covariance matrices must be diagonal).
gm_full = GaussianMixture(n_components=3, n_init=10, covariance_type="full", random_state=42)
gm_tied = GaussianMixture(n_components=3, n_init=10, covariance_type="tied", random_state=42)
gm_spherical = GaussianMixture(n_components=3, n_init=10, covariance_type="spherical", random_state=42)
gm_diag = GaussianMixture(n_components=3, n_init=10, covariance_type="diag", random_state=42)
gm_full.fit(X)
gm_tied.fit(X)
gm_spherical.fit(X)
gm_diag.fit(X)
def compare_gaussian_mixtures(gm1, gm2, X):
plt.figure(figsize=(9, 4))
plt.subplot(121)
plot_gaussian_mixture(gm1, X)
plt.title('covariance_type="{}"'.format(gm1.covariance_type), fontsize=14)
plt.subplot(122)
plot_gaussian_mixture(gm2, X, show_ylabels=False)
plt.title('covariance_type="{}"'.format(gm2.covariance_type), fontsize=14)
# +
compare_gaussian_mixtures(gm_tied, gm_spherical, X)
save_fig("covariance_type_plot")
plt.show()
# -
compare_gaussian_mixtures(gm_full, gm_diag, X)
plt.tight_layout()
plt.show()
# ## Anomaly Detection using Gaussian Mixtures
# Gaussian Mixtures can be used for _anomaly detection_: instances located in low-density regions can be considered anomalies. You must define what density threshold you want to use. For example, in a manufacturing company that tries to detect defective products, the ratio of defective products is usually well-known. Say it is equal to 4%, then you can set the density threshold to be the value that results in having 4% of the instances located in areas below that threshold density:
densities = gm.score_samples(X)
density_threshold = np.percentile(densities, 4)
anomalies = X[densities < density_threshold]
# +
plt.figure(figsize=(8, 4))
plot_gaussian_mixture(gm, X)
plt.scatter(anomalies[:, 0], anomalies[:, 1], color='r', marker='*')
plt.ylim(top=5.1)
save_fig("mixture_anomaly_detection_plot")
plt.show()
# -
# ## Model selection
# We cannot use the inertia or the silhouette score because they both assume that the clusters are spherical. Instead, we can try to find the model that minimizes a theoretical information criterion such as the Bayesian Information Criterion (BIC) or the Akaike Information Criterion (AIC):
#
# ${BIC} = {\log(m)p - 2\log({\hat L})}$
#
# ${AIC} = 2p - 2\log(\hat L)$
#
# * $m$ is the number of instances.
# * $p$ is the number of parameters learned by the model.
# * $\hat L$ is the maximized value of the likelihood function of the model. This is the conditional probability of the observed data $\mathbf{X}$, given the model and its optimized parameters.
#
# Both BIC and AIC penalize models that have more parameters to learn (e.g., more clusters), and reward models that fit the data well (i.e., models that give a high likelihood to the observed data).
gm.bic(X)
gm.aic(X)
# We could compute the BIC manually like this:
n_clusters = 3
n_dims = 2
n_params_for_weights = n_clusters - 1
n_params_for_means = n_clusters * n_dims
n_params_for_covariance = n_clusters * n_dims * (n_dims + 1) // 2
n_params = n_params_for_weights + n_params_for_means + n_params_for_covariance
max_log_likelihood = gm.score(X) * len(X) # log(L^)
bic = np.log(len(X)) * n_params - 2 * max_log_likelihood
aic = 2 * n_params - 2 * max_log_likelihood
bic, aic
n_params
# There's one weight per cluster, but the sum must be equal to 1, so we have one degree of freedom less, hence the -1. Similarly, the degrees of freedom for an $n \times n$ covariance matrix is not $n^2$, but $1 + 2 + \dots + n = \dfrac{n (n+1)}{2}$.
# Let's train Gaussian Mixture models with various values of $k$ and measure their BIC:
gms_per_k = [GaussianMixture(n_components=k, n_init=10, random_state=42).fit(X)
for k in range(1, 11)]
bics = [model.bic(X) for model in gms_per_k]
aics = [model.aic(X) for model in gms_per_k]
plt.figure(figsize=(8, 3))
plt.plot(range(1, 11), bics, "bo-", label="BIC")
plt.plot(range(1, 11), aics, "go--", label="AIC")
plt.xlabel("$k$", fontsize=14)
plt.ylabel("Information Criterion", fontsize=14)
plt.axis([1, 9.5, np.min(aics) - 50, np.max(aics) + 50])
plt.annotate('Minimum',
xy=(3, bics[2]),
xytext=(0.35, 0.6),
textcoords='figure fraction',
fontsize=14,
arrowprops=dict(facecolor='black', shrink=0.1)
)
plt.legend()
save_fig("aic_bic_vs_k_plot")
plt.show()
# Let's search for best combination of values for both the number of clusters and the `covariance_type` hyperparameter:
# +
min_bic = np.infty
for k in range(1, 11):
for covariance_type in ("full", "tied", "spherical", "diag"):
bic = GaussianMixture(n_components=k, n_init=10,
covariance_type=covariance_type,
random_state=42).fit(X).bic(X)
if bic < min_bic:
min_bic = bic
best_k = k
best_covariance_type = covariance_type
# -
best_k
best_covariance_type
# ## Variational Bayesian Gaussian Mixtures
# Rather than manually searching for the optimal number of clusters, it is possible to use instead the `BayesianGaussianMixture` class which is capable of giving weights equal (or close) to zero to unnecessary clusters. Just set the number of components to a value that you believe is greater than the optimal number of clusters, and the algorithm will eliminate the unnecessary clusters automatically.
from sklearn.mixture import BayesianGaussianMixture
bgm = BayesianGaussianMixture(n_components=10, n_init=10, random_state=42)
bgm.fit(X)
# The algorithm automatically detected that only 3 components are needed:
np.round(bgm.weights_, 2)
plt.figure(figsize=(8, 5))
plot_gaussian_mixture(bgm, X)
plt.show()
bgm_low = BayesianGaussianMixture(n_components=10, max_iter=1000, n_init=1,
weight_concentration_prior=0.01, random_state=42)
bgm_high = BayesianGaussianMixture(n_components=10, max_iter=1000, n_init=1,
weight_concentration_prior=10000, random_state=42)
nn = 73
bgm_low.fit(X[:nn])
bgm_high.fit(X[:nn])
np.round(bgm_low.weights_, 2)
np.round(bgm_high.weights_, 2)
# +
plt.figure(figsize=(9, 4))
plt.subplot(121)
plot_gaussian_mixture(bgm_low, X[:nn])
plt.title("weight_concentration_prior = 0.01", fontsize=14)
plt.subplot(122)
plot_gaussian_mixture(bgm_high, X[:nn], show_ylabels=False)
plt.title("weight_concentration_prior = 10000", fontsize=14)
save_fig("mixture_concentration_prior_plot")
plt.show()
# -
# Note: the fact that you see only 3 regions in the right plot although there are 4 centroids is not a bug. The weight of the top-right cluster is much larger than the weight of the lower-right cluster, so the probability that any given point in this region belongs to the top right cluster is greater than the probability that it belongs to the lower-right cluster.
X_moons, y_moons = make_moons(n_samples=1000, noise=0.05, random_state=42)
bgm = BayesianGaussianMixture(n_components=10, n_init=10, random_state=42)
bgm.fit(X_moons)
# +
plt.figure(figsize=(9, 3.2))
plt.subplot(121)
plot_data(X_moons)
plt.xlabel("$x_1$", fontsize=14)
plt.ylabel("$x_2$", fontsize=14, rotation=0)
plt.subplot(122)
plot_gaussian_mixture(bgm, X_moons, show_ylabels=False)
save_fig("moons_vs_bgm_plot")
plt.show()
# -
# Oops, not great... instead of detecting 2 moon-shaped clusters, the algorithm detected 8 ellipsoidal clusters. However, the density plot does not look too bad, so it might be usable for anomaly detection.
# ## Likelihood Function
from scipy.stats import norm
xx = np.linspace(-6, 4, 101)
ss = np.linspace(1, 2, 101)
XX, SS = np.meshgrid(xx, ss)
ZZ = 2 * norm.pdf(XX - 1.0, 0, SS) + norm.pdf(XX + 4.0, 0, SS)
ZZ = ZZ / ZZ.sum(axis=1)[:,np.newaxis] / (xx[1] - xx[0])
# +
from matplotlib.patches import Polygon
plt.figure(figsize=(8, 4.5))
x_idx = 85
s_idx = 30
plt.subplot(221)
plt.contourf(XX, SS, ZZ, cmap="GnBu")
plt.plot([-6, 4], [ss[s_idx], ss[s_idx]], "k-", linewidth=2)
plt.plot([xx[x_idx], xx[x_idx]], [1, 2], "b-", linewidth=2)
plt.xlabel(r"$x$")
plt.ylabel(r"$\theta$", fontsize=14, rotation=0)
plt.title(r"Model $f(x; \theta)$", fontsize=14)
plt.subplot(222)
plt.plot(ss, ZZ[:, x_idx], "b-")
max_idx = np.argmax(ZZ[:, x_idx])
max_val = np.max(ZZ[:, x_idx])
plt.plot(ss[max_idx], max_val, "r.")
plt.plot([ss[max_idx], ss[max_idx]], [0, max_val], "r:")
plt.plot([0, ss[max_idx]], [max_val, max_val], "r:")
plt.text(1.01, max_val + 0.005, r"$\hat{L}$", fontsize=14)
plt.text(ss[max_idx]+ 0.01, 0.055, r"$\hat{\theta}$", fontsize=14)
plt.text(ss[max_idx]+ 0.01, max_val - 0.012, r"$Max$", fontsize=12)
plt.axis([1, 2, 0.05, 0.15])
plt.xlabel(r"$\theta$", fontsize=14)
plt.grid(True)
plt.text(1.99, 0.135, r"$=f(x=2.5; \theta)$", fontsize=14, ha="right")
plt.title(r"Likelihood function $\mathcal{L}(\theta|x=2.5)$", fontsize=14)
plt.subplot(223)
plt.plot(xx, ZZ[s_idx], "k-")
plt.axis([-6, 4, 0, 0.25])
plt.xlabel(r"$x$", fontsize=14)
plt.grid(True)
plt.title(r"PDF $f(x; \theta=1.3)$", fontsize=14)
verts = [(xx[41], 0)] + list(zip(xx[41:81], ZZ[s_idx, 41:81])) + [(xx[80], 0)]
poly = Polygon(verts, facecolor='0.9', edgecolor='0.5')
plt.gca().add_patch(poly)
plt.subplot(224)
plt.plot(ss, np.log(ZZ[:, x_idx]), "b-")
max_idx = np.argmax(np.log(ZZ[:, x_idx]))
max_val = np.max(np.log(ZZ[:, x_idx]))
plt.plot(ss[max_idx], max_val, "r.")
plt.plot([ss[max_idx], ss[max_idx]], [-5, max_val], "r:")
plt.plot([0, ss[max_idx]], [max_val, max_val], "r:")
plt.axis([1, 2, -2.4, -2])
plt.xlabel(r"$\theta$", fontsize=14)
plt.text(ss[max_idx]+ 0.01, max_val - 0.05, r"$Max$", fontsize=12)
plt.text(ss[max_idx]+ 0.01, -2.39, r"$\hat{\theta}$", fontsize=14)
plt.text(1.01, max_val + 0.02, r"$\log \, \hat{L}$", fontsize=14)
plt.grid(True)
plt.title(r"$\log \, \mathcal{L}(\theta|x=2.5)$", fontsize=14)
save_fig("likelihood_function_plot")
plt.show()
# -
# # Exercise solutions
# ## 1. to 9.
# See Appendix A.
# ## 10. Cluster the Olivetti Faces Dataset
# *Exercise: The classic Olivetti faces dataset contains 400 grayscale 64 × 64–pixel images of faces. Each image is flattened to a 1D vector of size 4,096. 40 different people were photographed (10 times each), and the usual task is to train a model that can predict which person is represented in each picture. Load the dataset using the `sklearn.datasets.fetch_olivetti_faces()` function.*
# +
from sklearn.datasets import fetch_olivetti_faces
olivetti = fetch_olivetti_faces()
# -
print(olivetti.DESCR)
olivetti.target
# *Exercise: Then split it into a training set, a validation set, and a test set (note that the dataset is already scaled between 0 and 1). Since the dataset is quite small, you probably want to use stratified sampling to ensure that there are the same number of images per person in each set.*
# +
from sklearn.model_selection import StratifiedShuffleSplit
strat_split = StratifiedShuffleSplit(n_splits=1, test_size=40, random_state=42)
train_valid_idx, test_idx = next(strat_split.split(olivetti.data, olivetti.target))
X_train_valid = olivetti.data[train_valid_idx]
y_train_valid = olivetti.target[train_valid_idx]
X_test = olivetti.data[test_idx]
y_test = olivetti.target[test_idx]
strat_split = StratifiedShuffleSplit(n_splits=1, test_size=80, random_state=43)
train_idx, valid_idx = next(strat_split.split(X_train_valid, y_train_valid))
X_train = X_train_valid[train_idx]
y_train = y_train_valid[train_idx]
X_valid = X_train_valid[valid_idx]
y_valid = y_train_valid[valid_idx]
# -
print(X_train.shape, y_train.shape)
print(X_valid.shape, y_valid.shape)
print(X_test.shape, y_test.shape)
# To speed things up, we'll reduce the data's dimensionality using PCA:
# +
from sklearn.decomposition import PCA
pca = PCA(0.99)
X_train_pca = pca.fit_transform(X_train)
X_valid_pca = pca.transform(X_valid)
X_test_pca = pca.transform(X_test)
pca.n_components_
# -
# *Exercise: Next, cluster the images using K-Means, and ensure that you have a good number of clusters (using one of the techniques discussed in this chapter).*
# +
from sklearn.cluster import KMeans
k_range = range(5, 150, 5)
kmeans_per_k = []
for k in k_range:
print("k={}".format(k))
kmeans = KMeans(n_clusters=k, random_state=42).fit(X_train_pca)
kmeans_per_k.append(kmeans)
# +
from sklearn.metrics import silhouette_score
silhouette_scores = [silhouette_score(X_train_pca, model.labels_)
for model in kmeans_per_k]
best_index = np.argmax(silhouette_scores)
best_k = k_range[best_index]
best_score = silhouette_scores[best_index]
plt.figure(figsize=(8, 3))
plt.plot(k_range, silhouette_scores, "bo-")
plt.xlabel("$k$", fontsize=14)
plt.ylabel("Silhouette score", fontsize=14)
plt.plot(best_k, best_score, "rs")
plt.show()
# -
best_k
# It looks like the best number of clusters is quite high, at 120. You might have expected it to be 40, since there are 40 different people on the pictures. However, the same person may look quite different on different pictures (e.g., with or without glasses, or simply shifted left or right).
# +
inertias = [model.inertia_ for model in kmeans_per_k]
best_inertia = inertias[best_index]
plt.figure(figsize=(8, 3.5))
plt.plot(k_range, inertias, "bo-")
plt.xlabel("$k$", fontsize=14)
plt.ylabel("Inertia", fontsize=14)
plt.plot(best_k, best_inertia, "rs")
plt.show()
# -
# The optimal number of clusters is not clear on this inertia diagram, as there is no obvious elbow, so let's stick with k=120.
best_model = kmeans_per_k[best_index]
# *Exercise: Visualize the clusters: do you see similar faces in each cluster?*
# +
def plot_faces(faces, labels, n_cols=5):
n_rows = (len(faces) - 1) // n_cols + 1
plt.figure(figsize=(n_cols, n_rows * 1.1))
for index, (face, label) in enumerate(zip(faces, labels)):
plt.subplot(n_rows, n_cols, index + 1)
plt.imshow(face.reshape(64, 64), cmap="gray")
plt.axis("off")
plt.title(label)
plt.show()
for cluster_id in np.unique(best_model.labels_):
print("Cluster", cluster_id)
in_cluster = best_model.labels_==cluster_id
faces = X_train[in_cluster].reshape(-1, 64, 64)
labels = y_train[in_cluster]
plot_faces(faces, labels)
# -
# About 2 out of 3 clusters are useful: that is, they contain at least 2 pictures, all of the same person. However, the rest of the clusters have either one or more intruders, or they have just a single picture.
#
# Clustering images this way may be too imprecise to be directly useful when training a model (as we will see below), but it can be tremendously useful when labeling images in a new dataset: it will usually make labelling much faster.
# ## 11. Using Clustering as Preprocessing for Classification
# *Exercise: Continuing with the Olivetti faces dataset, train a classifier to predict which person is represented in each picture, and evaluate it on the validation set.*
# +
from sklearn.ensemble import RandomForestClassifier
clf = RandomForestClassifier(n_estimators=150, random_state=42)
clf.fit(X_train_pca, y_train)
clf.score(X_valid_pca, y_valid)
# -
# *Exercise: Next, use K-Means as a dimensionality reduction tool, and train a classifier on the reduced set.*
# +
X_train_reduced = best_model.transform(X_train_pca)
X_valid_reduced = best_model.transform(X_valid_pca)
X_test_reduced = best_model.transform(X_test_pca)
clf = RandomForestClassifier(n_estimators=150, random_state=42)
clf.fit(X_train_reduced, y_train)
clf.score(X_valid_reduced, y_valid)
# -
# Yikes! That's not better at all! Let's see if tuning the number of clusters helps.
# *Exercise: Search for the number of clusters that allows the classifier to get the best performance: what performance can you reach?*
# We could use a `GridSearchCV` like we did earlier in this notebook, but since we already have a validation set, we don't need K-fold cross-validation, and we're only exploring a single hyperparameter, so it's simpler to just run a loop manually:
# +
from sklearn.pipeline import Pipeline
for n_clusters in k_range:
pipeline = Pipeline([
("kmeans", KMeans(n_clusters=n_clusters, random_state=n_clusters)),
("forest_clf", RandomForestClassifier(n_estimators=150, random_state=42))
])
pipeline.fit(X_train_pca, y_train)
print(n_clusters, pipeline.score(X_valid_pca, y_valid))
# -
# Oh well, even by tuning the number of clusters, we never get beyond 80% accuracy. Looks like the distances to the cluster centroids are not as informative as the original images.
# *Exercise: What if you append the features from the reduced set to the original features (again, searching for the best number of clusters)?*
X_train_extended = np.c_[X_train_pca, X_train_reduced]
X_valid_extended = np.c_[X_valid_pca, X_valid_reduced]
X_test_extended = np.c_[X_test_pca, X_test_reduced]
clf = RandomForestClassifier(n_estimators=150, random_state=42)
clf.fit(X_train_extended, y_train)
clf.score(X_valid_extended, y_valid)
# That's a bit better, but still worse than without the cluster features. The clusters are not useful to directly train a classifier in this case (but they can still help when labelling new training instances).
# ## 12. A Gaussian Mixture Model for the Olivetti Faces Dataset
# *Exercise: Train a Gaussian mixture model on the Olivetti faces dataset. To speed up the algorithm, you should probably reduce the dataset's dimensionality (e.g., use PCA, preserving 99% of the variance).*
# +
from sklearn.mixture import GaussianMixture
gm = GaussianMixture(n_components=40, random_state=42)
y_pred = gm.fit_predict(X_train_pca)
# -
# *Exercise: Use the model to generate some new faces (using the `sample()` method), and visualize them (if you used PCA, you will need to use its `inverse_transform()` method).*
n_gen_faces = 20
gen_faces_reduced, y_gen_faces = gm.sample(n_samples=n_gen_faces)
gen_faces = pca.inverse_transform(gen_faces_reduced)
plot_faces(gen_faces, y_gen_faces)
# *Exercise: Try to modify some images (e.g., rotate, flip, darken) and see if the model can detect the anomalies (i.e., compare the output of the `score_samples()` method for normal images and for anomalies).*
# +
n_rotated = 4
rotated = np.transpose(X_train[:n_rotated].reshape(-1, 64, 64), axes=[0, 2, 1])
rotated = rotated.reshape(-1, 64*64)
y_rotated = y_train[:n_rotated]
n_flipped = 3
flipped = X_train[:n_flipped].reshape(-1, 64, 64)[:, ::-1]
flipped = flipped.reshape(-1, 64*64)
y_flipped = y_train[:n_flipped]
n_darkened = 3
darkened = X_train[:n_darkened].copy()
darkened[:, 1:-1] *= 0.3
darkened = darkened.reshape(-1, 64*64)
y_darkened = y_train[:n_darkened]
X_bad_faces = np.r_[rotated, flipped, darkened]
y_bad = np.concatenate([y_rotated, y_flipped, y_darkened])
plot_faces(X_bad_faces, y_bad)
# -
X_bad_faces_pca = pca.transform(X_bad_faces)
gm.score_samples(X_bad_faces_pca)
# The bad faces are all considered highly unlikely by the Gaussian Mixture model. Compare this to the scores of some training instances:
gm.score_samples(X_train_pca[:10])
# ## 13. Using Dimensionality Reduction Techniques for Anomaly Detection
# *Exercise: Some dimensionality reduction techniques can also be used for anomaly detection. For example, take the Olivetti faces dataset and reduce it with PCA, preserving 99% of the variance. Then compute the reconstruction error for each image. Next, take some of the modified images you built in the previous exercise, and look at their reconstruction error: notice how much larger the reconstruction error is. If you plot a reconstructed image, you will see why: it tries to reconstruct a normal face.*
# We already reduced the dataset using PCA earlier:
X_train_pca
def reconstruction_errors(pca, X):
X_pca = pca.transform(X)
X_reconstructed = pca.inverse_transform(X_pca)
mse = np.square(X_reconstructed - X).mean(axis=-1)
return mse
reconstruction_errors(pca, X_train).mean()
reconstruction_errors(pca, X_bad_faces).mean()
plot_faces(X_bad_faces, y_gen_faces)
X_bad_faces_reconstructed = pca.inverse_transform(X_bad_faces_pca)
plot_faces(X_bad_faces_reconstructed, y_gen_faces)
| 60,665 |
/notebooks/v1/28-ne-ne.ipynb | 76249f5b2eab78ba7db1021caf8e293118299b50 | [] | no_license | davidmcclure/mas-s61 | https://github.com/davidmcclure/mas-s61 | 0 | 0 | null | null | null | null | Jupyter Notebook | false | false | .py | 43,946 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: PUI2016_Python2
# language: python
# name: pui2016_python2
# ---
from __future__ import print_function
import pandas as pd
import numpy as np
import pylab as pl
# %pylab inline
import seaborn as sns
import statsmodels.api as st
firearms_df = pd.read_csv('World firearms murders and ownership - Sheet 1.csv')
guns_df = pd.read_csv('data-pvLFI.csv')
population_df = pd.read_csv('population.csv')
population_df.head(2)
population_2012_df = population_df[['Country Name','2012']].copy()
population_2012_df.head(2)
firearms_df.columns
guns_df.columns
firearms_df = firearms_df.merge(guns_df,left_on='Country/Territory',right_on='Country')
firearms_df.head(2)
firearms_df['color'] = "blue"
firearms_df.color[firearms_df['Country'] == 'United States'] = "red"
firearms_df = firearms_df.merge(population_2012_df,left_on='Country/Territory',right_on='Country Name')
firearms_df.columns
# ## plot the number of homicides by fire arm against the number of civilian firearms. plot the US in a different color
fig, ax = pl.subplots()
fig = figsize(12,8)
ax.scatter(firearms_df['Average total all civilian firearms'],firearms_df['Number of homicides by firearm'],
c=firearms_df['color'])
pl.xlabel("Avg Total Civilian Firearms")
pl.ylabel("Number of Homicides by Firearm")
pl.title("Civilian Firearms and Homicides")
# ### [Figure 1: Number of Civilian Firearms and Homicides]
fig, ax = pl.subplots()
fig = figsize(10,8)
ax.scatter(firearms_df['Average total all civilian firearms'],firearms_df['Number of homicides by firearm'],
c=firearms_df['color'])
e = pl.errorbar(firearms_df['Average total all civilian firearms'],
firearms_df['Number of homicides by firearm'],
yerr=np.sqrt(firearms_df['Number of homicides by firearm'] * 1.0),fmt='.',c='yellow',
label='error bar')
pl.xlabel("Avg Total Civilian Firearms")
pl.ylabel("Number of Homicides by Firearm")
pl.title("Civilian Firearms and Homicides")
pl.legend()
# ### [Figure 2: Number of Civilian Firearms and Homicides with error bars]
# ## plot the number of mass shootings against the number of civilian firearms. plot the US in a different color
fig, ax = pl.subplots()
fig = figsize(12,8)
ax.scatter(firearms_df['Average total all civilian firearms'],firearms_df['Number of mass shootings'],
c=firearms_df['color'])
pl.xlabel("Avg Total Civilian Firearms")
pl.ylabel("Number of mass shootings")
pl.title("Civilian Firearms and Mass Shootings")
# ### [Figure 3: Number of Civilian Firearms and Mass Shootings]
fig, ax = pl.subplots()
fig = figsize(12,8)
ax.scatter(firearms_df['Average total all civilian firearms'],firearms_df['Number of mass shootings'],
c=firearms_df['color'])
pl.errorbar(firearms_df['Average total all civilian firearms'],firearms_df['Number of mass shootings'],
yerr=np.sqrt(firearms_df['Number of mass shootings']*1.0),fmt='.',c='yellow',label='error bars')
pl.xlabel("Avg Total Civilian Firearms")
pl.ylabel("Number of mass shootings")
pl.title("Civilian Firearms and Mass Shootings")
pl.legend()
# ### [Figure 4: Number of Civilian Firearms and Mass Shootings with error bars]
# # Modeling
#
# ## fit a line to the Number of mass shootings per person as a function of Average total all civilian firearms per person.
mass_shooting_scaled = (firearms_df['Number of mass shootings']/firearms_df['2012'] * 1e3)/(1e-4)
civilian_firearams_scaled = firearms_df['Average total all civilian firearms']/firearms_df['2012']
pl.plot(civilian_firearams_scaled, mass_shooting_scaled,'o', label='y')
pl.xlabel('Avg Total Civilian Firearams')
pl.ylabel('Mass Shootings')
pl.legend()
model_ols = st.OLS(endog=mass_shooting_scaled, exog=civilian_firearams_scaled).fit()
model_ols.summary()
p1 = pl.plot(civilian_firearams_scaled, mass_shooting_scaled,'o',label='y')
p2 = pl.plot(civilian_firearams_scaled, model_ols.predict(),'b-',color='black',label='ols')
pl.xlabel('Avg Total Civilian Firearams')
pl.ylabel('Mass Shootings')
pl.title('Mass Shootings against Avg Total Civilian Firearms')
pl.legend()
# ### [Figure 5: Number of Civilian Firearms and Mass Shootings using OLS]
yerr = np.sqrt(mass_shooting_scaled.mean())
model_wls = st.WLS(endog=mass_shooting_scaled, exog=civilian_firearams_scaled, weights=1.0/yerr).fit()
model_wls.summary()
p1 = pl.plot(civilian_firearams_scaled, mass_shooting_scaled,'o',label='y')
p2 = pl.plot(civilian_firearams_scaled, model_ols.predict(),'b-',color='black',label='ols')
p3 = pl.plot(civilian_firearams_scaled, model_wls.predict(),'b-',color='red',label='wls')
pl.xlabel('Avg Total Civilian Firearams')
pl.ylabel('Mass Shootings')
pl.legend()
# ### [Figure 6: Number of Civilian Firearms and Mass Shootings using OLS and WLS]
df_polyfit = np.polyfit(civilian_firearams_scaled,mass_shooting_scaled,deg=2)
df_polyfit
# +
p1 = pl.plot(civilian_firearams_scaled, mass_shooting_scaled,'o',label='y')
pl.plot(civilian_firearams_scaled, civilian_firearams_scaled**2 * df_polyfit[0]+
civilian_firearams_scaled*df_polyfit[1]+df_polyfit[2],'b-',color='green',label='polyfit deg=2')
p2 = pl.plot(civilian_firearams_scaled, model_ols.predict(),'b-',color='black',label='ols')
pl.xlabel('Avg Total Civilian Firearams')
pl.ylabel('Mass Shootings')
pl.title('Number of civilian firearms and mass shootings')
pl.legend()
# -
# ### [Figure 7: Number of civilian firearms and mass shootings OLS and polyfit compared]
| 5,671 |
/Split.ipynb | 30e4e0f1f9ec4fe91c0d1124d7e4913064ee439a | [] | no_license | RodriguezRuizI/Split | https://github.com/RodriguezRuizI/Split | 0 | 0 | null | null | null | null | Jupyter Notebook | false | false | .py | 1,121 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
import pandas as pd
correos = pd.read_csv('lista-de-correos.csv')
correos['correo']=correos['Correos'].str.split('-', 1).str[1]
correos['correo'].to_csv('correos.csv')
# A veces splitea mal guiones de correos. Revisar manual
| 494 |
/book/building-tools/03-objects/introduction/.main.md.bcp.ipynb | 067d308ddc9b3522e493078ce122ffb4113e95b7 | [
"MIT"
] | permissive | daffidwilde/pfm | https://github.com/daffidwilde/pfm | 0 | 0 | MIT | 2020-10-28T14:39:03 | 2020-10-28T10:38:15 | null | Jupyter Notebook | false | false | .py | 1,191 | # ---
# jupyter:
# jupytext:
# formats: ipynb,md:myst
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Object oriented programming
#
# In the first part of this book we covered a number of tools that allow us to
# carry out mathematical techniques. One example of this is the `sympy.Symbol`
# object that create a symbolic variable that can be manipulated. In this chapter
# we will see how to define similar mathematical objects.
#
# ```{important}
# In this chapter we will cover:
#
# - Creating objects.
# - Giving objects attributes.
# - Defining methods on objects.
# - Inheriting new objects from others.
# ```
| 813 |
/Appendix-SGD.ipynb | 0d396aabebc329a99272e9c4f6997010c5d33151 | [] | no_license | Jackal08/neural-networks | https://github.com/Jackal08/neural-networks | 2 | 1 | null | 2017-06-25T16:42:24 | 2017-06-17T11:31:14 | null | Jupyter Notebook | false | false | .py | 48,257 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernel_info:
# name: dev
# kernelspec:
# display_name: Python [conda env:PythonAdv] *
# language: python
# name: conda-env-PythonAdv-py
# ---
# Update sklearn to prevent version mismatches
# !pip install sklearn --upgrade
# install joblib. This will be used to save your model.
# Restart your kernel after installing
# !pip install joblib
import pandas as pd
# # Read the CSV and Perform Basic Data Cleaning
df = pd.read_csv("Resources/exoplanet_data.csv")
# Drop the null columns where all values are null
df = df.dropna(axis='columns', how='all')
# Drop the null rows
df = df.dropna()
df.head()
# Checking the value count for the first column
df["koi_disposition"].value_counts()
# Filter out err columns
planets_filter_out_err = df.filter(regex = '^((?!err).)*$')
planets_filter_out_err.head()
# +
# Set the X and y
X = planets_filter_out_err.drop(columns = ["koi_disposition"])
y = planets_filter_out_err["koi_disposition"]
print(X.shape, y.shape)
# -
# # Create a Train Test Split
# +
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y, random_state=42, stratify =y)
# -
y.head()
# # Pre-processing
#
# Scale the data using the MinMaxScaler and perform some feature selection
from sklearn.preprocessing import LabelEncoder, MinMaxScaler
from tensorflow.keras.utils import to_categorical
# +
# Scale your data
X_scaler = MinMaxScaler().fit(X_train)
X_train_scaled = X_scaler.transform(X_train)
X_test_scaled = X_scaler.transform(X_test)
# Step 1: Label-encode data set
label_encoder = LabelEncoder()
label_encoder.fit(y_train)
encoded_y_train = label_encoder.transform(y_train)
encoded_y_test = label_encoder.transform(y_test)
# # Step 2: Convert encoded labels to one-hot-encoding
y_train_categorical = to_categorical(encoded_y_train)
y_test_categorical = to_categorical(encoded_y_test)
y_train_categorical
# -
y_train_categorical.shape
# # Train the Model
#
#
# +
from tensorflow.keras.models import Sequential
from tensorflow.keras.layers import Dense
model = Sequential()
model.add(Dense(units=100, activation='relu', input_dim=20))
model.add(Dense(units=3, activation='softmax'))
# -
# Compile the model
model.compile(optimizer='adam',
loss='categorical_crossentropy',
metrics=['accuracy'])
model.summary()
# +
from keras.callbacks import EarlyStopping
callbacks =[EarlyStopping('val_loss', patience=2)]
model.fit(
X_train_scaled,
y_train_categorical,
callbacks= callbacks,
epochs=150,
shuffle=True,
verbose=2
)
# -
# ## Deep Learning
deep_model = Sequential()
deep_model.add(Dense(units=100, activation='relu', input_dim=20))
deep_model.add(Dense(units=100, activation='relu'))
deep_model.add(Dense(units=3, activation='softmax'))
deep_model.summary()
# +
deep_model.compile(optimizer='adam',
loss='categorical_crossentropy',
metrics=['accuracy'])
deep_model.fit(
X_train_scaled,
y_train_categorical,
callbacks= callbacks,
epochs=150,
shuffle=True,
verbose=2
)
# -
# ## Compare the models below
model_loss, model_accuracy = model.evaluate(
X_test_scaled, y_test_categorical, verbose=2)
print(
f"Normal Neural Network - Loss: {model_loss}, Accuracy: {model_accuracy}")
model_loss, model_accuracy = deep_model.evaluate(
X_test_scaled, y_test_categorical, verbose=2)
print(f"Deep Neural Network - Loss: {model_loss}, Accuracy: {model_accuracy}")
# ## Saving Model
# Save the model
model.save("space_model_trained.h5")
# Save the model
deep_model.save("space_deep_model_trained.h5")
# Load the model
from tensorflow.keras.models import load_model
space_model = load_model("space_model_trained.h5")
space_deep_model = load_model("space_deep_model_trained.h5")
# ## Evaluating the loadedmodel
model_loss, model_accuracy = space_model.evaluate(
X_test_scaled, y_test_categorical, verbose=2)
print(
f"Normal Neural Network - Loss: {model_loss}, Accuracy: {model_accuracy}")
model_loss, model_accuracy = space_deep_model.evaluate(
X_test_scaled, y_test_categorical, verbose=2)
print(
f"Normal Neural Network - Loss: {model_loss}, Accuracy: {model_accuracy}")
Qlunc_yaml_inputs_1['Components']['Scanner']['Azimuth'][2]))))[0], # Focus distance in [meters]
cone_angle = np.tile(Qlunc_yaml_inputs_1['Components']['Scanner']['Cone angle'],(1,len(np.arange(Qlunc_yaml_inputs_1['Components']['Scanner']['Azimuth'][0],
Qlunc_yaml_inputs_1['Components']['Scanner']['Azimuth'][1],
Qlunc_yaml_inputs_1['Components']['Scanner']['Azimuth'][2]))))[0], # Cone angle in [degrees].
x = np.array(Qlunc_yaml_inputs_1['Components']['Scanner']['x']),
y = np.array(Qlunc_yaml_inputs_1['Components']['Scanner']['y']),
z = np.array(Qlunc_yaml_inputs_1['Components']['Scanner']['z']),
stdv_focus_dist = Qlunc_yaml_inputs_1['Components']['Scanner']['stdv focus distance'], # Focus distance standard deviation in [meters].
stdv_cone_angle = Qlunc_yaml_inputs_1['Components']['Scanner']['stdv Cone angle'], # Cone angle standard deviation in [degrees].
stdv_azimuth = Qlunc_yaml_inputs_1['Components']['Scanner']['stdv Azimuth'], # Azimuth angle standard deviation in [degrees].
unc_func = uopc.UQ_Scanner) #eval(Qlunc_yaml_inputs['Components']['Scanner']['Uncertainty function']) ) # here you put the function describing your scanner uncertainty.
Scanner2 = scanner(name = Qlunc_yaml_inputs_2['Components']['Scanner']['Name'], # Introduce your scanner name.
scanner_type = Qlunc_yaml_inputs_2['Components']['Scanner']['Type'],
origin = Qlunc_yaml_inputs_2['Components']['Scanner']['Origin'], # Origin (coordinates of the lidar deployment).
pattern = Qlunc_yaml_inputs_2['Components']['Scanner']['Pattern'],
sample_rate = Qlunc_yaml_inputs_2['Components']['Scanner']['Sample rate'], # for now introduce it in [degrees].
# This values for focus distance, cone_angle and azimuth define a typical VAD scanning sequence:
azimuth = np.array(np.arange(Qlunc_yaml_inputs_2['Components']['Scanner']['Azimuth'][0],
Qlunc_yaml_inputs_2['Components']['Scanner']['Azimuth'][1],
Qlunc_yaml_inputs_2['Components']['Scanner']['Azimuth'][2])), # Azimuth angle in [degrees].
focus_dist = np.tile(Qlunc_yaml_inputs_2['Components']['Scanner']['Focus distance'],(1,len(np.arange(Qlunc_yaml_inputs_2['Components']['Scanner']['Azimuth'][0],
Qlunc_yaml_inputs_2['Components']['Scanner']['Azimuth'][1],
Qlunc_yaml_inputs_2['Components']['Scanner']['Azimuth'][2]))))[0], # Focus distance in [meters]
cone_angle = np.tile(Qlunc_yaml_inputs_2['Components']['Scanner']['Cone angle'],(1,len(np.arange(Qlunc_yaml_inputs_2['Components']['Scanner']['Azimuth'][0],
Qlunc_yaml_inputs_2['Components']['Scanner']['Azimuth'][1],
Qlunc_yaml_inputs_2['Components']['Scanner']['Azimuth'][2]))))[0], # Cone angle in [degrees].
x = np.array(Qlunc_yaml_inputs_2['Components']['Scanner']['x']),
y = np.array(Qlunc_yaml_inputs_2['Components']['Scanner']['y']),
z = np.array(Qlunc_yaml_inputs_2['Components']['Scanner']['z']),
stdv_focus_dist = Qlunc_yaml_inputs_2['Components']['Scanner']['stdv focus distance'], # Focus distance standard deviation in [meters].
stdv_cone_angle = Qlunc_yaml_inputs_2['Components']['Scanner']['stdv Cone angle'], # Cone angle standard deviation in [degrees].
stdv_azimuth = Qlunc_yaml_inputs_2['Components']['Scanner']['stdv Azimuth'], # Azimuth angle standard deviation in [degrees].
unc_func = uopc.UQ_Scanner) #eval(Qlunc_yaml_inputs['Components']['Scanner']['Uncertainty function']) ) # here you put the function describing your scanner uncertainty.
# -
# ## Optical circulator:
# Once we have created the different scanning heads we can create the optical circulator. Since they use the same optical circulator we just have to create one for both lidars.
# +
#Optical Circulator:
Optical_circulator = optical_circulator (name = Qlunc_yaml_inputs_1['Components']['Optical Circulator']['Name'],
insertion_loss = Qlunc_yaml_inputs_1['Components']['Optical Circulator']['Insertion loss'],
unc_func = eval(Qlunc_yaml_inputs_1['Components']['Optical Circulator']['Uncertainty function']))
# -
# ## Optics modules:
# Create the optics modules by introducing scanners and optical circulator in them (by instantiating the optics `class`):
# +
# Optics Module lidar1:
Optics_Module = optics (name = Qlunc_yaml_inputs_1['Modules']['Optics Module']['Name'], # Introduce your Optics Module name.
scanner = eval(Qlunc_yaml_inputs_1['Modules']['Optics Module']['Scanner']), # Scanner instance (in this example "Scanner") or "None". "None" means that you don´t want to include Scanner in Optics Module, either in uncertainty calculations.
optical_circulator = eval(Qlunc_yaml_inputs_1['Modules']['Optics Module']['Optical circulator']), # Optical Circulator instance (in this example "Optical_circulator") or "None". "None" means that you don´t want to include Optical circulator in Optics Module, either in uncertainty calculations.
laser = eval(Qlunc_yaml_inputs_1['Modules']['Optics Module']['Laser']),
unc_func = eval(Qlunc_yaml_inputs_1['Modules']['Optics Module']['Uncertainty function']))
# Optics Module lidar2:
Optics_Module2 = optics (name = Qlunc_yaml_inputs_2['Modules']['Optics Module']['Name'], # Introduce your Optics Module name.
scanner = eval(Qlunc_yaml_inputs_2['Modules']['Optics Module']['Scanner']), # Scanner instance (in this example "Scanner") or "None". "None" means that you don´t want to include Scanner in Optics Module, either in uncertainty calculations.
optical_circulator = eval(Qlunc_yaml_inputs_2['Modules']['Optics Module']['Optical circulator']), # Optical Circulator instance (in this example "Optical_circulator") or "None". "None" means that you don´t want to include Optical circulator in Optics Module, either in uncertainty calculations.
laser = eval(Qlunc_yaml_inputs_2['Modules']['Optics Module']['Laser']),
unc_func = eval(Qlunc_yaml_inputs_2['Modules']['Optics Module']['Uncertainty function']))
# -
# ## Photonics:
# +
# Instantiating optical amplifier:
Optical_Amplifier = optical_amplifier(name = Qlunc_yaml_inputs_1['Components']['Optical Amplifier']['Name'], # Introduce your scanner name.
OA_NF = Qlunc_yaml_inputs_1['Components']['Optical Amplifier']['Optical amplifier noise figure'], # In [dB]. Can introduce it as a table from manufactures (in this example the data is taken from Thorlabs.com, in section EDFA\Graps) or introduce a single well-known value
OA_Gain = Qlunc_yaml_inputs_1['Components']['Optical Amplifier']['Optical amplifier gain'], # In [dB]. (in this example the data is taken from Thorlabs.com, in section EDFA\Specs)
unc_func = eval(Qlunc_yaml_inputs_1['Components']['Optical Amplifier']['Uncertainty function'])) # Function describing Optical Amplifier uncertainty. Further informaion in "UQ_Photonics_Classes.py" comments.
# -
# Photodetector digital twin:
# +
# Instantiating Photodetector:
Photodetector = photodetector(name = Qlunc_yaml_inputs_1['Components']['Photodetector']['Name'], # Introduce your photodetector name.
Photo_BandWidth = Qlunc_yaml_inputs_1['Components']['Photodetector']['Photodetector BandWidth'], # In[]. Photodetector bandwidth
Load_Resistor = Qlunc_yaml_inputs_1['Components']['Photodetector']['Load resistor'], # In [ohms]
Photo_efficiency = Qlunc_yaml_inputs_1['Components']['Photodetector']['Photodetector efficiency'], # Photodetector efficiency [-]
Dark_Current = Qlunc_yaml_inputs_1['Components']['Photodetector']['Dark current'], # In [A]. Dark current in the photodetector.
Photo_SignalP = Qlunc_yaml_inputs_1['Components']['Photodetector']['Photodetector signalP'],
Power_interval = np.array(np.arange(Qlunc_yaml_inputs_1['Components']['Photodetector']['Power interval'][0],
Qlunc_yaml_inputs_1['Components']['Photodetector']['Power interval'][1],
Qlunc_yaml_inputs_1['Components']['Photodetector']['Power interval'][2])),#np.arange(Qlunc_yaml_inputs['Components']['Photodetector']['Power interval']), # In [w]. Power interval for the photodetector domain in photodetector SNR plot.
Gain_TIA = Qlunc_yaml_inputs_1['Components']['Photodetector']['Gain TIA'], # In [dB]. If there is a transimpedance amplifier.
V_Noise_TIA = Qlunc_yaml_inputs_1['Components']['Photodetector']['V Noise TIA'], # In [V]. If there is a transimpedance amplifier.
unc_func = eval(Qlunc_yaml_inputs_1['Components']['Photodetector']['Uncertainty function'])) # Function describing Photodetector uncertainty. Further informaion in "UQ_Photonics_Classes.py" comments.
# -
# Photonics module digital twin:
# And finally the Photonics module:
#
# +
# Instantiating Photonics module:
Photonics_Module = photonics(name = Qlunc_yaml_inputs_1['Modules']['Photonics Module']['Name'], # Introduce your Photonics module name
photodetector = eval(Qlunc_yaml_inputs_1['Modules']['Photonics Module']['Photodetector']), # Photodetector instance (in this example "Photodetector") or "None". "None" means that you don´t want to include photodetector in Photonics Module, either in uncertainty calculations.
optical_amplifier = eval(Qlunc_yaml_inputs_1['Modules']['Photonics Module']['Optical amplifier']), # Scanner instance (in this example "OpticalAmplifier") or "None". "None" means that you don´t want to include Optical Amplifier in Photonics Module, either in uncertainty calculations.
unc_func = eval(Qlunc_yaml_inputs_1['Modules']['Photonics Module']['Uncertainty function']))
# -
# Lidar general inputs:
# +
# Instantiating lidar general inputs
Lidar_inputs = lidar_gral_inp(name = Qlunc_yaml_inputs_1['Components']['Lidar general inputs']['Name'],
wave = Qlunc_yaml_inputs_1['Components']['Lidar general inputs']['Wavelength'],
yaw_error = Qlunc_yaml_inputs_1['Components']['Lidar general inputs']['Yaw error'],
pitch_error = Qlunc_yaml_inputs_1['Components']['Lidar general inputs']['Pitch error'],
roll_error = Qlunc_yaml_inputs_1['Components']['Lidar general inputs']['Roll error'])
Lidar_inputs_2 = lidar_gral_inp(name = Qlunc_yaml_inputs_2['Components']['Lidar general inputs']['Name'],
wave = Qlunc_yaml_inputs_2['Components']['Lidar general inputs']['Wavelength'],
yaw_error = Qlunc_yaml_inputs_2['Components']['Lidar general inputs']['Yaw error'],
pitch_error = Qlunc_yaml_inputs_2['Components']['Lidar general inputs']['Pitch error'],
roll_error = Qlunc_yaml_inputs_2['Components']['Lidar general inputs']['Roll error'])
# -
# Atmospheric scenarios:
#
# We can create different atmospheric scenarios under which the lidar uncertainties are assessed. Thus, single or multiple scenarios can be evaluated either including single or time-seried atmospheric variables values.
# +
# Instantiating atmospheric scenario(s): Assuming both lidars ahve the same atmospheric
Atmospheric_TimeSeries = Qlunc_yaml_inputs_1['Atmospheric_inputs']['TimeSeries']
if Atmospheric_TimeSeries:
Atmos_TS_FILE = '../metadata/AtmosphericData/'+Qlunc_yaml_inputs_1['Atmospheric_inputs']['Atmos_TS_FILE']
AtmosphericScenarios_TS = pd.read_csv(Atmos_TS_FILE,delimiter=';',decimal=',')
Atmospheric_inputs = {
'temperature' : list(AtmosphericScenarios_TS.loc[:,'T']),
'humidity' : list(AtmosphericScenarios_TS.loc[:,'H']),
'rain' : list(AtmosphericScenarios_TS.loc[:,'rain']),
'fog' : list(AtmosphericScenarios_TS.loc[:,'fog']),
'time' : list(AtmosphericScenarios_TS.loc[:,'t'])
}
Atmospheric_Scenario = atmosphere(name = 'Atmosphere1',
temperature = Atmospheric_inputs['temperature'])
else:
Atmospheric_Scenario = atmosphere(name = 'Atmosphere1',
temperature = Qlunc_yaml_inputs_1['Atmospheric_inputs']['Temperature'])
# -
# Now we build up the lidar devices by putting all components together:
# +
Lidar_1 = lidar(name = Qlunc_yaml_inputs_1['Lidar']['Name'], # Introduce the name of your lidar device.
photonics = eval(Qlunc_yaml_inputs_1['Lidar']['Photonics module']), # Introduce the name of your photonics module.
optics = eval(Qlunc_yaml_inputs_1['Lidar']['Optics module']), # Introduce the name of your optics module.
power = eval(Qlunc_yaml_inputs_1['Lidar']['Power module']), # Introduce the name of your power module. NOT IMPLEMENTED YET!
lidar_inputs = eval(Qlunc_yaml_inputs_1['Lidar']['Lidar inputs']), # Introduce lidar general inputs
unc_func = eval(Qlunc_yaml_inputs_1['Lidar']['Uncertainty function'])) # Function estimating lidar global uncertainty
Lidar_2 = lidar(name = Qlunc_yaml_inputs_2['Lidar']['Name'], # Introduce the name of your lidar device.
photonics = eval(Qlunc_yaml_inputs_2['Lidar']['Photonics module']), # Introduce the name of your photonics module.
optics = eval(Qlunc_yaml_inputs_2['Lidar']['Optics module']), # Introduce the name of your optics module.
power = eval(Qlunc_yaml_inputs_2['Lidar']['Power module']), # Introduce the name of your power module. NOT IMPLEMENTED YET!
lidar_inputs = eval(Qlunc_yaml_inputs_2['Lidar']['Lidar inputs']), # Introduce lidar general inputs
unc_func = eval(Qlunc_yaml_inputs_2['Lidar']['Uncertainty function'])) # Function estimating lidar global uncertainty
# -
# and plot both lidars together:
# Plotting parameters:
plot_param={
'axes_label_fontsize' : 16,
'textbox_fontsize' : 14,
'title_fontsize' : 18,
'suptitle_fontsize' : 23,
'legend_fontsize' : 12,
'xlim' : [-25,25],
'ylim' : [-25,25],
'zlim' : [0,90],
'markersize' : 5,
'markersize_lidar' : 9,
'marker' : '.',
'markerTheo' : '.b',
'tick_labelrotation' : 45,
'Qlunc_version' : 'Qlunc Version - 0.9'
}
# +
# Calling Scanner uncertainty to plot the graphics
Scanner_Data1 = Lidar_1.optics.scanner.Uncertainty(Lidar_1,Atmospheric_Scenario,cts,Qlunc_yaml_inputs_1)
Scanner_Data2 = Lidar_2.optics.scanner.Uncertainty(Lidar_2,Atmospheric_Scenario,cts,Qlunc_yaml_inputs_2)
# Creating the figure and the axes
fig,axs4 = plt.subplots()
axs4=plt.axes(projection='3d')
# Plotting
# First lidar (Lidar_1)
axs4.plot([Lidar_1.optics.scanner.origin[0]],[Lidar_1.optics.scanner.origin[1]],[Lidar_1.optics.scanner.origin[2]],'ob',label='{} coordinates [{},{},{}]'.format(Lidar_1.LidarID,Lidar_1.optics.scanner.origin[0],Lidar_1.optics.scanner.origin[1],Lidar_1.optics.scanner.origin[2]),markersize=plot_param['markersize_lidar'])
axs4.plot(Scanner_Data1['MeasPoint_Coordinates'][0],Scanner_Data1['MeasPoint_Coordinates'][1],Scanner_Data1['MeasPoint_Coordinates'][2],plot_param['markerTheo'],markersize=plot_param['markersize'],label='Theoretical measuring point')
axs4.plot(Scanner_Data1['NoisyMeasPoint_Coordinates'][0],Scanner_Data1['NoisyMeasPoint_Coordinates'][1],Scanner_Data1['NoisyMeasPoint_Coordinates'][2],plot_param['marker'],markersize=plot_param['markersize'],label='Distance error [m] = {0:.3g}$\pm${1:.3g}'.format(np.mean(Scanner_Data1['Simu_Mean_Distance']),np.mean(Scanner_Data1['STDV_Distance'])))
# Second lidar(Lidar_2)
axs4.plot([Lidar_2.optics.scanner.origin[0]],[Lidar_2.optics.scanner.origin[1]],[Lidar_2.optics.scanner.origin[2]],'ob',label='{} coordinates [{},{},{}]'.format(Lidar_2.LidarID,Lidar_2.optics.scanner.origin[0],Lidar_2.optics.scanner.origin[1],Lidar_2.optics.scanner.origin[2]),markersize=plot_param['markersize_lidar'])
axs4.plot(Scanner_Data2['MeasPoint_Coordinates'][0],Scanner_Data2['MeasPoint_Coordinates'][1],Scanner_Data2['MeasPoint_Coordinates'][2],plot_param['markerTheo'],markersize=plot_param['markersize'],label='Theoretical measuring point')
axs4.plot(Scanner_Data2['NoisyMeasPoint_Coordinates'][0],Scanner_Data2['NoisyMeasPoint_Coordinates'][1],Scanner_Data2['NoisyMeasPoint_Coordinates'][2],plot_param['marker'],markersize=plot_param['markersize'],label='Distance error [m] = {0:.3g}$\pm${1:.3g}'.format(np.mean(Scanner_Data2['Simu_Mean_Distance']),np.mean(Scanner_Data2['STDV_Distance'])))
# Setting labels, legend, title and axes limits:
axs4.set_xlabel('x [m]',fontsize=plot_param['axes_label_fontsize'])#,orientation=plot_param['tick_labelrotation'])
axs4.set_ylabel('y [m]',fontsize=plot_param['axes_label_fontsize'])#,orientation=plot_param['tick_labelrotation'])
axs4.set_zlabel('z [m]',fontsize=plot_param['axes_label_fontsize'])
axs4.set_title('Scanner Pointing accuracy',fontsize=plot_param['title_fontsize'])
# axs4.legend()
axs4.set_xlim3d(plot_param['xlim'][0],plot_param['xlim'][1])
axs4.set_ylim3d(plot_param['ylim'][0],plot_param['ylim'][1])
axs4.set_zlim3d(plot_param['zlim'][0],plot_param['zlim'][1])
# -
| 25,001 |
/Image_Classification_with_CNNs.ipynb | e815b4058ea3cd8556110961dda13c8d2bc024b6 | [] | no_license | abhiwalia15/Fashion-MNIST | https://github.com/abhiwalia15/Fashion-MNIST | 0 | 0 | null | null | null | null | Jupyter Notebook | false | false | .py | 35,012 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Python 3
# name: python3
# ---
# + [markdown] id="view-in-github" colab_type="text"
# <a href="https://colab.research.google.com/github/landges/automatic-octo-machine/blob/master/cipfa_and_hips.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# + [markdown] id="wQuocPO60hA6" colab_type="text"
# Установочные команды
# + id="kheVlN2kUDg4" colab_type="code" colab={}
# !pip install tokenization
# !pip install nltk
# !pip install pymorphy2
# + [markdown] id="KjddqDSZ0qCY" colab_type="text"
# Импорты и используемые функции
# + id="tgWrqpib9ZrY" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 34} outputId="3feae512-72bf-4a35-eebc-89ea92e85dd5"
from google.colab import drive
import re
import tokenization
import nltk
from nltk import sent_tokenize, word_tokenize, regexp_tokenize
from nltk.corpus import stopwords
import pymorphy2
import matplotlib.pyplot as plt
import os
drive.mount('/content/drive')
def normalize_tokens(tokens):
morph = pymorphy2.MorphAnalyzer()
return [morph.parse(tok)[0].normal_form for tok in tokens]
def get_normalize_text(text):
text=text.lower()
words=re.findall(r"[\w']+|[.,!?;:{}() \n]",text)
words=normalize_tokens(words)
words=[t for t in words if len(t)>2]
return words
def uniq_dic(words):
vocab={}
for word in words:
vocab[word]=vocab.get(word,0)+1
return vocab
def hips(n,k=100,b=0.6):
return k*(n**b)
def sort_text(text):
return len(text)
# + [markdown] id="U9qOjIpi0szg" colab_type="text"
# ***ГРАФИЧКИ***
# + [markdown] id="9QOzO2jJvgWc" colab_type="text"
#
#
# > [Закон ЦИПФА](https://ru.wikipedia.org/wiki/%D0%97%D0%B0%D0%BA%D0%BE%D0%BD_%D0%A6%D0%B8%D0%BF%D1%84%D0%B0)
#
# Зако́н Ци́пфа («ранг—частота») — эмпирическая закономерность распределения частоты слов естественного языка: если все слова языка (или просто достаточно длинного текста) упорядочить по убыванию частоты их использования, то частота n-го слова в таком списке окажется приблизительно обратно пропорциональной его порядковому номеру n
#
#
#
# + id="pgfaoP7uxOW8" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 312} outputId="b0f84fd2-108e-478b-ef3a-71535f2d01ef"
s=open('/content/drive/My Drive/Colab Notebooks/тексты/35000Толстой Лев. Война и мир. Том 3 - royallib.ru.txt','r')
text=s.read()
s.close()
words=get_normalize_text(text)
vocab=uniq_dic(words)
# print(*sorted(sorted(vocab), key=lambda x :vocab.get(x),reverse=False), sep ='\n')
list_id=list(vocab.items())
list_id.sort(key=lambda i :i[1],reverse=True)
id=[t[1] for t in list_id]
plt.plot(range(100,len(id)),id[100:])
plt.title("Закон ЦИПФА")
plt.xlabel("Range")
plt.ylabel("Frequency")
# + [markdown] id="FeJfegIlvku9" colab_type="text"
#
#
# > [Закон ХИПСА](https://ru.wikipedia.org/wiki/%D0%97%D0%B0%D0%BA%D0%BE%D0%BD_%D0%A5%D0%B8%D0%BF%D1%81%D0%B0)
#
# Зако́н Хи́пса — эмпирическая закономерность в лингвистике, описывающая распределение числа разных слов в документе (или наборе документов) как функцию от его длины. Описывается формулой
#
#
# 
#
# где VR — число разных слов в тексте размера n. K и β — свободные параметры, определяются эмпирически. Для английского корпуса текстов, K обычно лежит между 10 и 100, а β между 0.4 и 0.6.
#
# + id="D0E1FuqIoNNP" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 312} outputId="00385d17-108c-4844-c29c-c2c2425092b9"
texts=[]
directory='/content/drive/My Drive/Colab Notebooks/тексты/'
files = os.listdir(directory)
for file in files:
s=open(directory+file,'r')
texts.append(s.read())
s.close
texts.sort(key=sort_text)
f=[len(uniq_dic(get_normalize_text(r))) for r in texts]
len_text=[len(t) for t in texts]
plt.plot(len_text,f)
plt.title("Закон ХИПСА")
plt.xlabel("length of text")
plt.ylabel("count of uniqueness words")
of clothing at low resolution (28 $\times$ 28 pixels), as seen here:
#
# <table>
# <tr><td>
# <img src="https://tensorflow.org/images/fashion-mnist-sprite.png"
# alt="Fashion MNIST sprite" width="600">
# </td></tr>
# <tr><td align="center">
# <b>Figure 1.</b> <a href="https://github.com/zalandoresearch/fashion-mnist">Fashion-MNIST samples</a> (by Zalando, MIT License).<br/>
# </td></tr>
# </table>
#
# Fashion MNIST is intended as a drop-in replacement for the classic [MNIST](http://yann.lecun.com/exdb/mnist/) dataset—often used as the "Hello, World" of machine learning programs for computer vision. The MNIST dataset contains images of handwritten digits (0, 1, 2, etc) in an identical format to the articles of clothing we'll use here.
#
# This guide uses Fashion MNIST for variety, and because it's a slightly more challenging problem than regular MNIST. Both datasets are relatively small and are used to verify that an algorithm works as expected. They're good starting points to test and debug code.
#
# We will use 60,000 images to train the network and 10,000 images to evaluate how accurately the network learned to classify images. You can access the Fashion MNIST directly from TensorFlow, using the [Datasets](https://www.tensorflow.org/datasets) API:
# + colab_type="code" id="7MqDQO0KCaWS" colab={}
dataset, metadata = tfds.load('fashion_mnist', as_supervised=True, with_info=True)
train_dataset, test_dataset = dataset['train'], dataset['test']
# + [markdown] colab_type="text" id="t9FDsUlxCaWW"
# Loading the dataset returns metadata as well as a *training dataset* and *test dataset*.
#
# * The model is trained using `train_dataset`.
# * The model is tested against `test_dataset`.
#
# The images are 28 $\times$ 28 arrays, with pixel values in the range `[0, 255]`. The *labels* are an array of integers, in the range `[0, 9]`. These correspond to the *class* of clothing the image represents:
#
# <table>
# <tr>
# <th>Label</th>
# <th>Class</th>
# </tr>
# <tr>
# <td>0</td>
# <td>T-shirt/top</td>
# </tr>
# <tr>
# <td>1</td>
# <td>Trouser</td>
# </tr>
# <tr>
# <td>2</td>
# <td>Pullover</td>
# </tr>
# <tr>
# <td>3</td>
# <td>Dress</td>
# </tr>
# <tr>
# <td>4</td>
# <td>Coat</td>
# </tr>
# <tr>
# <td>5</td>
# <td>Sandal</td>
# </tr>
# <tr>
# <td>6</td>
# <td>Shirt</td>
# </tr>
# <tr>
# <td>7</td>
# <td>Sneaker</td>
# </tr>
# <tr>
# <td>8</td>
# <td>Bag</td>
# </tr>
# <tr>
# <td>9</td>
# <td>Ankle boot</td>
# </tr>
# </table>
#
# Each image is mapped to a single label. Since the *class names* are not included with the dataset, store them here to use later when plotting the images:
# + colab_type="code" id="IjnLH5S2CaWx" colab={}
class_names = ['T-shirt/top', 'Trouser', 'Pullover', 'Dress', 'Coat',
'Sandal', 'Shirt', 'Sneaker', 'Bag', 'Ankle boot']
# + [markdown] colab_type="text" id="Brm0b_KACaWX"
# ### Explore the data
#
# Let's explore the format of the dataset before training the model. The following shows there are 60,000 images in the training set, and 10000 images in the test set:
# + colab_type="code" id="MaOTZxFzi48X" colab={}
num_train_examples = metadata.splits['train'].num_examples
num_test_examples = metadata.splits['test'].num_examples
print("Number of training examples: {}".format(num_train_examples))
print("Number of test examples: {}".format(num_test_examples))
# + [markdown] colab_type="text" id="ES6uQoLKCaWr"
# ## Preprocess the data
#
# The value of each pixel in the image data is an integer in the range `[0,255]`. For the model to work properly, these values need to be normalized to the range `[0,1]`. So here we create a normalization function, and then apply it to each image in the test and train datasets.
# + colab_type="code" id="nAsH3Zm-76pB" colab={}
def normalize(images, labels):
images = tf.cast(images, tf.float32)
images /= 255
return images, labels
# The map function applies the normalize function to each element in the train
# and test datasets
train_dataset = train_dataset.map(normalize)
test_dataset = test_dataset.map(normalize)
# + [markdown] colab_type="text" id="lIQbEiJGXM-q"
# ### Explore the processed data
#
# Let's plot an image to see what it looks like.
# + colab_type="code" id="oSzE9l7PjHx0" colab={}
# Take a single image, and remove the color dimension by reshaping
for image, label in test_dataset.take(1):
break
image = image.numpy().reshape((28,28))
# Plot the image - voila a piece of fashion clothing
plt.figure()
plt.imshow(image, cmap=plt.cm.binary)
plt.colorbar()
plt.grid(False)
plt.show()
# + [markdown] colab_type="text" id="Ee638AlnCaWz"
# Display the first 25 images from the *training set* and display the class name below each image. Verify that the data is in the correct format and we're ready to build and train the network.
# + colab_type="code" id="oZTImqg_CaW1" colab={}
plt.figure(figsize=(10,10))
i = 0
for (image, label) in test_dataset.take(25):
image = image.numpy().reshape((28,28))
plt.subplot(5,5,i+1)
plt.xticks([])
plt.yticks([])
plt.grid(False)
plt.imshow(image, cmap=plt.cm.binary)
plt.xlabel(class_names[label])
i += 1
plt.show()
# + [markdown] colab_type="text" id="59veuiEZCaW4"
# ## Build the model
#
# Building the neural network requires configuring the layers of the model, then compiling the model.
# + [markdown] colab_type="text" id="Gxg1XGm0eOBy"
# ### Setup the layers
#
# The basic building block of a neural network is the *layer*. A layer extracts a representation from the data fed into it. Hopefully, a series of connected layers results in a representation that is meaningful for the problem at hand.
#
# Much of deep learning consists of chaining together simple layers. Most layers, like `tf.keras.layers.Dense`, have internal parameters which are adjusted ("learned") during training.
# + colab_type="code" id="9ODch-OFCaW4" colab={}
model = tf.keras.Sequential([
tf.keras.layers.Conv2D(32, (3,3), padding='same', activation=tf.nn.relu,
input_shape=(28, 28, 1)),
tf.keras.layers.MaxPooling2D((2, 2), strides=2),
tf.keras.layers.Conv2D(64, (3,3), padding='same', activation=tf.nn.relu),
tf.keras.layers.MaxPooling2D((2, 2), strides=2),
tf.keras.layers.Flatten(),
tf.keras.layers.Dense(128, activation=tf.nn.relu),
tf.keras.layers.Dense(10, activation=tf.nn.softmax)
])
# + [markdown] colab_type="text" id="gut8A_7rCaW6"
# This network layers are:
#
# * **"convolutions"** `tf.keras.layers.Conv2D and MaxPooling2D`— Network start with two pairs of Conv/MaxPool. The first layer is a Conv2D filters (3,3) being applied to the input image, retaining the original image size by using padding, and creating 32 output (convoluted) images (so this layer creates 32 convoluted images of the same size as input). After that, the 32 outputs are reduced in size using a MaxPooling2D (2,2) with a stride of 2. The next Conv2D also has a (3,3) kernel, takes the 32 images as input and creates 64 outputs which are again reduced in size by a MaxPooling2D layer. So far in the course, we have described what a Convolution does, but we haven't yet covered how you chain multiples of these together. We will get back to this in lesson 4 when we use color images. At this point, it's enough if you understand the kind of operation a convolutional filter performs
#
# * **output** `tf.keras.layers.Dense` — A 128-neuron, followed by 10-node *softmax* layer. Each node represents a class of clothing. As in the previous layer, the final layer takes input from the 128 nodes in the layer before it, and outputs a value in the range `[0, 1]`, representing the probability that the image belongs to that class. The sum of all 10 node values is 1.
#
#
# ### Compile the model
#
# Before the model is ready for training, it needs a few more settings. These are added during the model's *compile* step:
#
#
# * *Loss function* — An algorithm for measuring how far the model's outputs are from the desired output. The goal of training is this measures loss.
# * *Optimizer* —An algorithm for adjusting the inner parameters of the model in order to minimize loss.
# * *Metrics* —Used to monitor the training and testing steps. The following example uses *accuracy*, the fraction of the images that are correctly classified.
# + colab_type="code" id="Lhan11blCaW7" colab={}
model.compile(optimizer='adam',
loss='sparse_categorical_crossentropy',
metrics=['accuracy'])
# + [markdown] colab_type="text" id="qKF6uW-BCaW-"
# ## Train the model
#
# First, we define the iteration behavior for the train dataset:
# 1. Repeat forever by specifying `dataset.repeat()` (the `epochs` parameter described below limits how long we perform training).
# 2. The `dataset.shuffle(60000)` randomizes the order so our model cannot learn anything from the order of the examples.
# 3. And `dataset.batch(32)` tells `model.fit` to use batches of 32 images and labels when updating the model variables.
#
# Training is performed by calling the `model.fit` method:
# 1. Feed the training data to the model using `train_dataset`.
# 2. The model learns to associate images and labels.
# 3. The `epochs=5` parameter limits training to 5 full iterations of the training dataset, so a total of 5 * 60000 = 300000 examples.
#
# (Don't worry about `steps_per_epoch`, the requirement to have this flag will soon be removed.)
# + colab_type="code" id="o_Dp8971McQ1" colab={}
BATCH_SIZE = 32
train_dataset = train_dataset.repeat().shuffle(num_train_examples).batch(BATCH_SIZE)
test_dataset = test_dataset.batch(BATCH_SIZE)
# + colab_type="code" id="xvwvpA64CaW_" colab={}
model.fit(train_dataset, epochs=10, steps_per_epoch=math.ceil(num_train_examples/BATCH_SIZE))
# + [markdown] colab_type="text" id="W3ZVOhugCaXA"
# As the model trains, the loss and accuracy metrics are displayed. This model reaches an accuracy of about 0.97 (or 97%) on the training data.
# + [markdown] colab_type="text" id="oEw4bZgGCaXB"
# ## Evaluate accuracy
#
# Next, compare how the model performs on the test dataset. Use all examples we have in the test dataset to assess accuracy.
# + colab_type="code" id="VflXLEeECaXC" colab={}
test_loss, test_accuracy = model.evaluate(test_dataset, steps=math.ceil(num_test_examples/32))
print('Accuracy on test dataset:', test_accuracy)
# + [markdown] colab_type="text" id="yWfgsmVXCaXG"
# As it turns out, the accuracy on the test dataset is smaller than the accuracy on the training dataset. This is completely normal, since the model was trained on the `train_dataset`. When the model sees images it has never seen during training, (that is, from the `test_dataset`), we can expect performance to go down.
# + [markdown] colab_type="text" id="xsoS7CPDCaXH"
# ## Make predictions and explore
#
# With the model trained, we can use it to make predictions about some images.
# + colab_type="code" id="Ccoz4conNCpl" colab={}
for test_images, test_labels in test_dataset.take(1):
test_images = test_images.numpy()
test_labels = test_labels.numpy()
predictions = model.predict(test_images)
# + colab_type="code" id="Gl91RPhdCaXI" colab={}
predictions.shape
# + [markdown] colab_type="text" id="x9Kk1voUCaXJ"
# Here, the model has predicted the label for each image in the testing set. Let's take a look at the first prediction:
# + colab_type="code" id="3DmJEUinCaXK" colab={}
predictions[0]
# + [markdown] colab_type="text" id="-hw1hgeSCaXN"
# A prediction is an array of 10 numbers. These describe the "confidence" of the model that the image corresponds to each of the 10 different articles of clothing. We can see which label has the highest confidence value:
# + colab_type="code" id="qsqenuPnCaXO" colab={}
np.argmax(predictions[0])
# + [markdown] colab_type="text" id="E51yS7iCCaXO"
# So the model is most confident that this image is a shirt, or `class_names[6]`. And we can check the test label to see this is correct:
# + colab_type="code" id="Sd7Pgsu6CaXP" colab={}
test_labels[0]
# + [markdown] colab_type="text" id="ygh2yYC972ne"
# We can graph this to look at the full set of 10 channels
# + colab_type="code" id="DvYmmrpIy6Y1" colab={}
def plot_image(i, predictions_array, true_labels, images):
predictions_array, true_label, img = predictions_array[i], true_labels[i], images[i]
plt.grid(False)
plt.xticks([])
plt.yticks([])
plt.imshow(img[...,0], cmap=plt.cm.binary)
predicted_label = np.argmax(predictions_array)
if predicted_label == true_label:
color = 'blue'
else:
color = 'red'
plt.xlabel("{} {:2.0f}% ({})".format(class_names[predicted_label],
100*np.max(predictions_array),
class_names[true_label]),
color=color)
def plot_value_array(i, predictions_array, true_label):
predictions_array, true_label = predictions_array[i], true_label[i]
plt.grid(False)
plt.xticks([])
plt.yticks([])
thisplot = plt.bar(range(10), predictions_array, color="#777777")
plt.ylim([0, 1])
predicted_label = np.argmax(predictions_array)
thisplot[predicted_label].set_color('red')
thisplot[true_label].set_color('blue')
# + [markdown] colab_type="text" id="d4Ov9OFDMmOD"
# Let's look at the 0th image, predictions, and prediction array.
# + colab_type="code" id="HV5jw-5HwSmO" colab={}
i = 0
plt.figure(figsize=(6,3))
plt.subplot(1,2,1)
plot_image(i, predictions, test_labels, test_images)
plt.subplot(1,2,2)
plot_value_array(i, predictions, test_labels)
# + colab_type="code" id="Ko-uzOufSCSe" colab={}
i = 12
plt.figure(figsize=(6,3))
plt.subplot(1,2,1)
plot_image(i, predictions, test_labels, test_images)
plt.subplot(1,2,2)
plot_value_array(i, predictions, test_labels)
# + [markdown] colab_type="text" id="kgdvGD52CaXR"
# Let's plot several images with their predictions. Correct prediction labels are blue and incorrect prediction labels are red. The number gives the percent (out of 100) for the predicted label. Note that it can be wrong even when very confident.
# + colab_type="code" id="hQlnbqaw2Qu_" colab={}
# Plot the first X test images, their predicted label, and the true label
# Color correct predictions in blue, incorrect predictions in red
num_rows = 5
num_cols = 3
num_images = num_rows*num_cols
plt.figure(figsize=(2*2*num_cols, 2*num_rows))
for i in range(num_images):
plt.subplot(num_rows, 2*num_cols, 2*i+1)
plot_image(i, predictions, test_labels, test_images)
plt.subplot(num_rows, 2*num_cols, 2*i+2)
plot_value_array(i, predictions, test_labels)
# + [markdown] colab_type="text" id="R32zteKHCaXT"
# Finally, use the trained model to make a prediction about a single image.
# + colab_type="code" id="yRJ7JU7JCaXT" colab={}
# Grab an image from the test dataset
img = test_images[0]
print(img.shape)
# + [markdown] colab_type="text" id="vz3bVp21CaXV"
# `tf.keras` models are optimized to make predictions on a *batch*, or collection, of examples at once. So even though we're using a single image, we need to add it to a list:
# + colab_type="code" id="lDFh5yF_CaXW" colab={}
# Add the image to a batch where it's the only member.
img = np.array([img])
print(img.shape)
# + [markdown] colab_type="text" id="EQ5wLTkcCaXY"
# Now predict the image:
# + colab_type="code" id="o_rzNSdrCaXY" colab={}
predictions_single = model.predict(img)
print(predictions_single)
# + colab_type="code" id="6Ai-cpLjO-3A" colab={}
plot_value_array(0, predictions_single, test_labels)
_ = plt.xticks(range(10), class_names, rotation=45)
# + [markdown] colab_type="text" id="cU1Y2OAMCaXb"
# `model.predict` returns a list of lists, one for each image in the batch of data. Grab the predictions for our (only) image in the batch:
# + colab_type="code" id="2tRmdq_8CaXb" colab={}
np.argmax(predictions_single[0])
# + [markdown] colab_type="text" id="YFc2HbEVCaXd"
# And, as before, the model predicts a label of 6 (shirt).
# + [markdown] colab_type="text" id="-KtnHECKZni_"
# # Exercises
#
# Experiment with different models and see how the accuracy results differ. In particular change the following parameters:
# * Set training epochs set to 1
# * Number of neurons in the Dense layer following the Flatten one. For example, go really low (e.g. 10) in ranges up to 512 and see how accuracy changes
# * Add additional Dense layers between the Flatten and the final Dense(10, activation=tf.nn.softmax), experiment with different units in these layers
# * Don't normalize the pixel values, and see the effect that has
#
#
# Remember to enable GPU to make everything run faster (Runtime -> Change runtime type -> Hardware accelerator -> GPU).
# Also, if you run into trouble, simply reset the entire environment and start from the beginning:
# * Edit -> Clear all outputs
# * Runtime -> Reset all runtimes
# + colab_type="code" id="WIIYx5IIfwF0" colab={}
| 21,255 |
/C3_W1_Assignment_Bird_Boxes.ipynb | 3d2045d41c0cbf4bcf687ad6add148dfbe24ebba | [] | no_license | seonokkim/Advanced-Computer-Vision-with-TensorFlow | https://github.com/seonokkim/Advanced-Computer-Vision-with-TensorFlow | 0 | 0 | null | null | null | null | Jupyter Notebook | false | false | .py | 880,547 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# + [markdown] id="view-in-github" colab_type="text"
# <a href="https://colab.research.google.com/github/sokim0991/Advanced-Computer-Vision-with-TensorFlow/blob/main/C3_W1_Assignment_Bird_Boxes.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# + [markdown] id="KsjDCIat6_UK"
# # Predicting Bounding Boxes
#
# Welcome to Course 3, Week 1 Programming Assignment!
#
# In this week's assignment, you'll build a model to predict bounding boxes around images.
# - You will use transfer learning on any of the pre-trained models available in Keras.
# - You'll be using the [Caltech Birds - 2010](http://www.vision.caltech.edu/visipedia/CUB-200.html) dataset.
#
#
# ### How to submit your work
# Notice that there is not a "submit assignment" button in this notebook.
#
# To check your work and get graded on your work, you'll train the model, save it and then upload the model to Coursera for grading.
# + [markdown] id="8iA95a8EKXR5"
# - [Initial steps](#0)
# - [0.1 Set up your Colab](#0-1)
# - [0.2 Set up the data location](#0-2)
# - [0.3 Choose the GPU Runtime](#0-3)
# - [0.4 Mount your drive](#0-4)
# - [0.5 Imports](#0-5)
# - [1. Visualization Utilities](#1)
# - [1.1 Bounding Boxes Utilities](#1-1)
# - [1.2 Data and Predictions Utilities](#1-2)
# - [2. Preprocessing and Loading the Dataset](#2)
# - [2.1 Preprocessing Utilities](#2-1)
# - [2.2 Visualize the prepared Data](#2-2)
# - [2.3 Loading the Dataset](#2-3)
# - [3. Define the Network](#3)
# - [Exercise 1](#ex-01)
# - [Exercise 2](#ex-02)
# - [Exercise 3](#ex-03)
# - [Exercise 4](#ex-04)
# - [Exercise 5](#ex-05)
# - [4. Training the Model](#4)
# - [Prepare to train the model](#4.1)
# - [Exercise 6](#ex-06)
# - [Fit the model to the data](#4.2)
# - [Exercise 7](#ex-07)
# - [5. Validate the Model](#5)
# - [5.1 Loss](#5-1)
# - [5.2 Save your Model](#5-2)
# - [5.3 Plot the Loss Function](#5-3)
# - [5.4 Evaluate performance using IoU](#5-4)
# - [6. Visualize Predictions](#6)
# - [7. Upload your model for grading](#7)
#
# + [markdown] id="19sIQEkEKXR5"
# <a name="0"></a>
# ## 0. Initial steps
# + [markdown] id="EQXm4O39KXR5"
# <a name="0-1"></a>
# ## 0.1 Set up your Colab
#
# - As you cannot save the changes you make to this colab, you have to make a copy of this notebook in your own drive and run that.
# - You can do so by going to `File -> Save a copy in Drive`.
# - Close this colab and open the copy which you have made in your own drive. Then continue to the next step to set up the data location.
# + [markdown] id="khsTrAlcKXR5"
# <a name="0-2"></a>
# ## Set up the data location
# A copy of the dataset that you'll be using is stored in a publicly viewable Google Drive folder. You'll want to add a shortcut to it to your own Google Drive.
# - Go to this google drive folder named [TF3 C3 W1 Data](https://drive.google.com/drive/folders/1xgqUw9uWzL5Kh88iPdX1TBQgnkc-wVKd?usp=sharing)
# - Next to the folder name "TF3 C3 W1 Data", hover your mouse over the triangle to reveal the drop down menu.
# - Use the drop down menu to select `"Add shortcut to Drive"` A pop-up menu will open up.
# - In the pop-up menu, "My Drive" is selected by default. Click the `ADD SHORTCUT` button. This should add a shortcut to the folder `TF3 C3 W1 Data` within your own google drive at the location `content/drive`.
# - To verify, go to the left-side menu and click on "My Drive". Scroll through your files to look for the shortcut TF3 C3 W1 Data.
#
# Please make sure this happens, as you'll be reading the data for this notebook from this folder.
# + id="e6adWu1Be3-7" colab={"base_uri": "https://localhost:8080/"} outputId="fa5dd278-45b0-4374-a894-7accef866f7f"
from google.colab import drive
drive.mount('/content/drive')
# + [markdown] id="Wz0OAuIEKXR5"
# <a name="0-3"></a>
# ## 0.3 Choose the GPU Runtime
# - Make sure your runtime is **GPU** (_not_ CPU or TPU). And if it is an option, make sure you are using _Python 3_. You can select these settings by going to `Runtime -> Change runtime type -> Select the above mentioned settings and then press SAVE`
# + [markdown] id="tgJOkF1VKXR5"
# <a name="0-4"></a>
# ## 0.4 Mount your drive
#
# Please run the next code cell and follow these steps to mount your Google Drive so that it can be accessed by this Colab.
# - Run the code cell below. A web link will appear below the cell.
# - Please click on the web link, which will open a new tab in your browser, which asks you to choose your google account.
# - Choose your google account to login.
# - The page will display "Google Drive File Stream wants to access your Google Account". Please click "Allow".
# - The page will now show a code (a line of text). Please copy the code and return to this Colab.
# - Paste the code the textbox that is labeled "Enter your authorization code:" and hit `<Enter>`
# - The text will now say "Mounted at /content/drive/"
# + id="Q6Pnix2iKXR5" colab={"base_uri": "https://localhost:8080/"} outputId="4181346b-bbf8-4f94-d104-11c4c8d0b34b"
from google.colab import drive
drive.mount('/content/drive/', force_remount=True)
# + [markdown] id="qpiJj8ym0v0-"
# <a name="0-5"></a>
# ## 0.5 Imports
# + id="DRl07kRr7uny" colab={"base_uri": "https://localhost:8080/"} outputId="304f8cc9-4db2-41b3-cdf5-421994841145"
# If you get a checksum error with the dataset, you'll need this
# !pip install tfds-nightly==4.0.1.dev202010100107
# + id="AoilhmYe1b5t"
import os, re, time, json
import PIL.Image, PIL.ImageFont, PIL.ImageDraw
import numpy as np
import tensorflow as tf
from matplotlib import pyplot as plt
import tensorflow_datasets as tfds
import cv2
# + [markdown] id="ddcggt0WKXR5"
# Store the path to the data.
# - Remember to follow the steps to `set up the data location` (above) so that you'll have a shortcut to the data in your Google Drive.
# + id="upfxqqK0vTMc"
data_dir = "/content/drive/My Drive/TF3 C3 W1 Data/"
# + [markdown] id="xmoFKEd98MP3"
# <a name="1"></a>
# ## 1. Visualization Utilities
#
# + [markdown] id="WOhS3mNlDOLX"
# <a name="1-1"></a>
# ### 1.1 Bounding Boxes Utilities
#
# We have provided you with some functions which you will use to draw bounding boxes around the birds in the `image`.
#
# - `draw_bounding_box_on_image`: Draws a single bounding box on an image.
# - `draw_bounding_boxes_on_image`: Draws multiple bounding boxes on an image.
# - `draw_bounding_boxes_on_image_array`: Draws multiple bounding boxes on an array of images.
# + id="YWIHFPa0uOC_"
def draw_bounding_box_on_image(image, ymin, xmin, ymax, xmax, color=(255, 0, 0), thickness=5):
"""
Adds a bounding box to an image.
Bounding box coordinates can be specified in either absolute (pixel) or
normalized coordinates by setting the use_normalized_coordinates argument.
Args:
image: a PIL.Image object.
ymin: ymin of bounding box.
xmin: xmin of bounding box.
ymax: ymax of bounding box.
xmax: xmax of bounding box.
color: color to draw bounding box. Default is red.
thickness: line thickness. Default value is 4.
"""
image_width = image.shape[1]
image_height = image.shape[0]
cv2.rectangle(image, (int(xmin), int(ymin)), (int(xmax), int(ymax)), color, thickness)
def draw_bounding_boxes_on_image(image, boxes, color=[], thickness=5):
"""
Draws bounding boxes on image.
Args:
image: a PIL.Image object.
boxes: a 2 dimensional numpy array of [N, 4]: (ymin, xmin, ymax, xmax).
The coordinates are in normalized format between [0, 1].
color: color to draw bounding box. Default is red.
thickness: line thickness. Default value is 4.
Raises:
ValueError: if boxes is not a [N, 4] array
"""
boxes_shape = boxes.shape
if not boxes_shape:
return
if len(boxes_shape) != 2 or boxes_shape[1] != 4:
raise ValueError('Input must be of size [N, 4]')
for i in range(boxes_shape[0]):
draw_bounding_box_on_image(image, boxes[i, 1], boxes[i, 0], boxes[i, 3],
boxes[i, 2], color[i], thickness)
def draw_bounding_boxes_on_image_array(image, boxes, color=[], thickness=5):
"""
Draws bounding boxes on image (numpy array).
Args:
image: a numpy array object.
boxes: a 2 dimensional numpy array of [N, 4]: (ymin, xmin, ymax, xmax).
The coordinates are in normalized format between [0, 1].
color: color to draw bounding box. Default is red.
thickness: line thickness. Default value is 4.
display_str_list_list: a list of strings for each bounding box.
Raises:
ValueError: if boxes is not a [N, 4] array
"""
draw_bounding_boxes_on_image(image, boxes, color, thickness)
return image
# + [markdown] id="USx9tRBF8hWy"
# <a name="1-2"></a>
# ### 1.2 Data and Predictions Utilities
#
# We've given you some helper functions and code that are used to visualize the data and the model's predictions.
#
# - `display_digits_with_boxes`: This displays a row of "digit" images along with the model's predictions for each image.
# - `plot_metrics`: This plots a given metric (like loss) as it changes over multiple epochs of training.
# + id="nwJ4rZ1d_7ql"
# Matplotlib config
plt.rc('image', cmap='gray')
plt.rc('grid', linewidth=0)
plt.rc('xtick', top=False, bottom=False, labelsize='large')
plt.rc('ytick', left=False, right=False, labelsize='large')
plt.rc('axes', facecolor='F8F8F8', titlesize="large", edgecolor='white')
plt.rc('text', color='a8151a')
plt.rc('figure', facecolor='F0F0F0')# Matplotlib fonts
MATPLOTLIB_FONT_DIR = os.path.join(os.path.dirname(plt.__file__), "mpl-data/fonts/ttf")
# utility to display a row of digits with their predictions
def display_digits_with_boxes(images, pred_bboxes, bboxes, iou, title, bboxes_normalized=False):
n = len(images)
fig = plt.figure(figsize=(20, 4))
plt.title(title)
plt.yticks([])
plt.xticks([])
for i in range(n):
ax = fig.add_subplot(1, 10, i+1)
bboxes_to_plot = []
if (len(pred_bboxes) > i):
bbox = pred_bboxes[i]
bbox = [bbox[0] * images[i].shape[1], bbox[1] * images[i].shape[0], bbox[2] * images[i].shape[1], bbox[3] * images[i].shape[0]]
bboxes_to_plot.append(bbox)
if (len(bboxes) > i):
bbox = bboxes[i]
if bboxes_normalized == True:
bbox = [bbox[0] * images[i].shape[1],bbox[1] * images[i].shape[0], bbox[2] * images[i].shape[1], bbox[3] * images[i].shape[0] ]
bboxes_to_plot.append(bbox)
img_to_draw = draw_bounding_boxes_on_image_array(image=images[i], boxes=np.asarray(bboxes_to_plot), color=[(255,0,0), (0, 255, 0)])
plt.xticks([])
plt.yticks([])
plt.imshow(img_to_draw)
if len(iou) > i :
color = "black"
if (iou[i][0] < iou_threshold):
color = "red"
ax.text(0.2, -0.3, "iou: %s" %(iou[i][0]), color=color, transform=ax.transAxes)
# utility to display training and validation curves
def plot_metrics(metric_name, title, ylim=5):
plt.title(title)
plt.ylim(0,ylim)
plt.plot(history.history[metric_name],color='blue',label=metric_name)
plt.plot(history.history['val_' + metric_name],color='green',label='val_' + metric_name)
# + [markdown] id="JVkc7nzg-WUy"
# <a name="2"></a>
# ## 2. Preprocess and Load the Dataset
# + [markdown] id="4Xv_8MbApX23"
# <a name="2-1"></a>
# ### 2.1 Preprocessing Utilities
#
# We have given you some helper functions to pre-process the image data.
# + [markdown] id="dg66jHMvw_f8"
# #### read_image_tfds
# - Resizes `image` to (224, 224)
# - Normalizes `image`
# - Translates and normalizes bounding boxes
# + id="MEEyTpmNxS0A"
def read_image_tfds(image, bbox):
image = tf.cast(image, tf.float32)
shape = tf.shape(image)
factor_x = tf.cast(shape[1], tf.float32)
factor_y = tf.cast(shape[0], tf.float32)
image = tf.image.resize(image, (224, 224,))
image = image/127.5
image -= 1
bbox_list = [bbox[0] / factor_x ,
bbox[1] / factor_y,
bbox[2] / factor_x ,
bbox[3] / factor_y]
return image, bbox_list
# + [markdown] id="zxqvA3wkyH7p"
# #### read_image_with_shape
# This is very similar to `read_image_tfds` except it also keeps a copy of the original image (before pre-processing) and returns this as well.
# - Makes a copy of the original image.
# - Resizes `image` to (224, 224)
# - Normalizes `image`
# - Translates and normalizes bounding boxes
# + id="f10wa31DyeQ4"
def read_image_with_shape(image, bbox):
original_image = image
image, bbox_list = read_image_tfds(image, bbox)
return original_image, image, bbox_list
# + [markdown] id="oNEpxvyLykzo"
# #### read_image_tfds_with_original_bbox
#
# - This function reads `image` from `data`
# - It also denormalizes the bounding boxes (it undoes the bounding box normalization that is performed by the previous two helper functions.)
# + id="gsQo9vvhyoKb"
def read_image_tfds_with_original_bbox(data):
image = data["image"]
bbox = data["bbox"]
shape = tf.shape(image)
factor_x = tf.cast(shape[1], tf.float32)
factor_y = tf.cast(shape[0], tf.float32)
bbox_list = [bbox[1] * factor_x ,
bbox[0] * factor_y,
bbox[3] * factor_x,
bbox[2] * factor_y]
return image, bbox_list
# + [markdown] id="2ElJ9VX0yui9"
# #### dataset_to_numpy_util
# This function converts a `dataset` into numpy arrays of images and boxes.
# - This will be used when visualizing the images and their bounding boxes
# + id="CF-luxkJyzIA"
def dataset_to_numpy_util(dataset, batch_size=0, N=0):
# eager execution: loop through datasets normally
take_dataset = dataset.shuffle(1024)
if batch_size > 0:
take_dataset = take_dataset.batch(batch_size)
if N > 0:
take_dataset = take_dataset.take(N)
if tf.executing_eagerly():
ds_images, ds_bboxes = [], []
for images, bboxes in take_dataset:
ds_images.append(images.numpy())
ds_bboxes.append(bboxes.numpy())
return (np.array(ds_images), np.array(ds_bboxes))
# + [markdown] id="JZSf8zvBy2RX"
# #### dataset_to_numpy_with_original_bboxes_util
#
# - This function converts a `dataset` into numpy arrays of
# - original images
# - resized and normalized images
# - bounding boxes
# - This will be used for plotting the original images with true and predicted bounding boxes.
# + id="ZE8dgyPC1_6m"
def dataset_to_numpy_with_original_bboxes_util(dataset, batch_size=0, N=0):
normalized_dataset = dataset.map(read_image_with_shape)
if batch_size > 0:
normalized_dataset = normalized_dataset.batch(batch_size)
if N > 0:
normalized_dataset = normalized_dataset.take(N)
if tf.executing_eagerly():
ds_original_images, ds_images, ds_bboxes = [], [], []
for original_images, images, bboxes in normalized_dataset:
ds_images.append(images.numpy())
ds_bboxes.append(bboxes.numpy())
ds_original_images.append(original_images.numpy())
return np.array(ds_original_images), np.array(ds_images), np.array(ds_bboxes)
# + [markdown] id="I4gB0hprzMw4"
# <a name="2-2"></a>
# ### 2.2 Visualize the images and their bounding box labels
# Now you'll take a random sample of images from the training and validation sets and visualize them by plotting the corresponding bounding boxes.
# + [markdown] id="XUPENeUHKXR6"
# Visualize the **training** images and their bounding box labels
# + id="HW_AyCNIKXR6" colab={"base_uri": "https://localhost:8080/", "height": 818} outputId="46752a9c-0ece-40c7-f78b-bf1a2c194b6c"
def get_visualization_training_dataset():
dataset, info = tfds.load("caltech_birds2010", split="train", with_info=True, data_dir=data_dir)
print(info)
visualization_training_dataset = dataset.map(read_image_tfds_with_original_bbox,
num_parallel_calls=16)
return visualization_training_dataset
visualization_training_dataset = get_visualization_training_dataset()
(visualization_training_images, visualization_training_bboxes) = dataset_to_numpy_util(visualization_training_dataset, N=10)
display_digits_with_boxes(np.array(visualization_training_images), np.array([]), np.array(visualization_training_bboxes), np.array([]), "training images and their bboxes")
# + [markdown] id="2qCuoUtYKXR6"
# Visualize the **validation** images and their bounding boxes
# + id="XLGiEyK_KXR6" colab={"base_uri": "https://localhost:8080/", "height": 282} outputId="1c534928-22bb-4d60-d382-b557a195e7b5"
def get_visualization_validation_dataset():
dataset = tfds.load("caltech_birds2010", split="test", try_gcs=True, data_dir=data_dir)
visualization_validation_dataset = dataset.map(read_image_tfds_with_original_bbox, num_parallel_calls=16)
return visualization_validation_dataset
visualization_validation_dataset = get_visualization_validation_dataset()
(visualization_validation_images, visualization_validation_bboxes) = dataset_to_numpy_util(visualization_validation_dataset, N=10)
display_digits_with_boxes(np.array(visualization_validation_images), np.array([]), np.array(visualization_validation_bboxes), np.array([]), "validation images and their bboxes")
# + [markdown] id="h2f2DWcnzZRq"
# <a name="2-3"></a>
# ### 2.3 Load and prepare the datasets for the model
#
# These next two functions read and prepare the datasets that you'll feed to the model.
# - They use `read_image_tfds` to resize, and normalize each image and its bounding box label.
# - They performs shuffling and batching.
# - You'll use these functions to create `training_dataset` and `validation_dataset`, which you will give to the model that you're about to build.
# + id="5shayI_tzdq0"
BATCH_SIZE = 64
def get_training_dataset(dataset):
dataset = dataset.map(read_image_tfds, num_parallel_calls=16)
dataset = dataset.shuffle(512, reshuffle_each_iteration=True)
dataset = dataset.repeat()
dataset = dataset.batch(BATCH_SIZE)
dataset = dataset.prefetch(-1)
return dataset
def get_validation_dataset(dataset):
dataset = dataset.map(read_image_tfds, num_parallel_calls=16)
dataset = dataset.batch(BATCH_SIZE)
dataset = dataset.repeat()
return dataset
training_dataset = get_training_dataset(visualization_training_dataset)
validation_dataset = get_validation_dataset(visualization_validation_dataset)
# + [markdown] id="f8nHWWkS_eeZ"
# <a name="3"></a>
# ## 3. Define the Network
#
# Bounding box prediction is treated as a "regression" task, in that you want the model to output numerical values.
#
# - You will be performing transfer learning with **MobileNet V2**. The model architecture is available in TensorFlow Keras.
# - You'll also use pretrained `'imagenet'` weights as a starting point for further training. These weights are also readily available
# - You will choose to retrain all layers of **MobileNet V2** along with the final classification layers.
#
# **Note:** For the following exercises, please use the TensorFlow Keras Functional API (as opposed to the Sequential API).
# + [markdown] id="csyBuMZReYON"
# <a name='ex-01'></a>
# ### Exercise 1
#
# Please build a feature extractor using MobileNetV2.
#
# - First, create an instance of the mobilenet version 2 model
# - Please check out the documentation for [MobileNetV2](https://www.tensorflow.org/api_docs/python/tf/keras/applications/MobileNetV2)
# - Set the following parameters:
# - input_shape: (height, width, channel): input images have height and width of 224 by 224, and have red, green and blue channels.
# - include_top: you do not want to keep the "top" fully connected layer, since you will customize your model for the current task.
# - weights: Use the pre-trained 'imagenet' weights.
#
# - Next, make the feature extractor for your specific inputs by passing the `inputs` into your mobilenet model.
# - For example, if you created a model object called `some_model` and have inputs stored in `x`, you'd invoke the model and pass in your inputs like this: `some_model(x)` to get the feature extractor for your given inputs `x`.
#
# **Note**: please use mobilenet_v2 and not mobile_net or mobile_net_v3
# + id="7DFecRhe0Pqc"
'''
def feature_extractor(inputs):
### YOUR CODE HERE ###
# Create a mobilenet version 2 model object
mobilenet_model = None
# pass the inputs into this modle object to get a feature extractor for these inputs
feature_extractor = None
### END CODE HERE ###
# return the feature_extractor
return feature_extractor
'''
def feature_extractor(inputs):
### YOUR CODE HERE ###
# Create a mobilenet version 2 model object
mobilenet_model = tf.keras.applications.MobileNetV2(input_shape=(224, 224, 3),
include_top=False,
weights='imagenet')
# pass the inputs into this modle object to get a feature extractor for these inputs
feature_extractor = mobilenet_model(inputs)
### END CODE HERE ###
# return the feature_extractor
return feature_extractor
# + [markdown] id="4ufMK9Qy0VPM"
# <a name='ex-02'></a>
# ### Exercise 2
#
# Next, you'll define the dense layers to be used by your model.
#
# You'll be using the following layers
# - [GlobalAveragePooling2D](https://www.tensorflow.org/api_docs/python/tf/keras/layers/GlobalAveragePooling2D): pools the `features`.
# - [Flatten](https://www.tensorflow.org/api_docs/python/tf/keras/layers/Flatten): flattens the pooled layer.
# - [Dense](https://www.tensorflow.org/api_docs/python/tf/keras/layers/Dense): Add two dense layers:
# - A dense layer with 1024 neurons and a relu activation.
# - A dense layer following that with 512 neurons and a relu activation.
#
# **Note**: Remember, please build the model using the Functional API syntax (as opposed to the Sequential API).
# + id="0njchQxB0b4Q"
'''
def dense_layers(features):
### YOUR CODE HERE ###
# global average pooling 2D layer.
x = None
# flatten layer
x = None
# 1024 Dense layer, with relu
x = None
# 512 Dense layer, with relu
x = None
### END CODE HERE ###
return x
'''
def dense_layers(features):
### YOUR CODE HERE ###
# global average pooling 2D layer.
x = tf.keras.layers.GlobalAveragePooling2D()(features)
# flatten layer
x = tf.keras.layers.Flatten()(x)
# 1024 Dense layer, with relu
x = tf.keras.layers.Dense(1024, activation='relu')(x)
# 512 Dense layer, with relu
x = tf.keras.layers.Dense(512, activation='relu')(x)
### END CODE HERE ###
return x
# + [markdown] id="g7ARvWYw0sje"
# <a name='ex-03'></a>
# ### Exercise 3
#
#
# Now you'll define a layer that outputs the bounding box predictions.
# - You'll use a [Dense](https://www.tensorflow.org/api_docs/python/tf/keras/layers/Dense) layer.
# - Remember that you have _4 units_ in the output layer, corresponding to (xmin, ymin, xmax, ymax).
# - The prediction layer follows the previous dense layer, which is passed into this function as the variable `x`.
# - For grading purposes, please set the `name` parameter of this Dense layer to be `bounding_box'
# + id="VdsD0-Jl07zW"
'''
def bounding_box_regression(x):
### YOUR CODE HERE ###
# Dense layer named `bounding_box`
bounding_box_regression_output = None
### END CODE HERE ###
return bounding_box_regression_output
'''
def bounding_box_regression(x):
### YOUR CODE HERE ###
# Dense layer named `bounding_box`
bounding_box_regression_output = tf.keras.layers.Dense(units = '4', name = 'bounding_box')(x)
### END CODE HERE ###
return bounding_box_regression_output
# + [markdown] id="ELxJoKqu1OnM"
# <a name='ex-04'></a>
# ### Exercise 4
#
# Now, you'll use those functions that you have just defined above to construct the model.
# - feature_extractor(inputs)
# - dense_layers(features)
# - bounding_box_regression(x)
#
# Then you'll define the model object using [Model](https://www.tensorflow.org/s/results?q=Model). Set the two parameters:
# - inputs
# - outputs
# + id="wn9O9c7I1XRJ"
'''
def final_model(inputs):
### YOUR CODE HERE ###
# features
feature_cnn = None
# dense layers
last_dense_layer = None
# bounding box
bounding_box_output = None
# define the TensorFlow Keras model using the inputs and outputs to your model
model = None
### END CODE HERE ###
return model
'''
def final_model(inputs):
### YOUR CODE HERE ###
# features
feature_cnn = feature_extractor(inputs)
# dense layers
last_dense_layer = dense_layers(feature_cnn)
# bounding box
bounding_box_output = bounding_box_regression(last_dense_layer)
# define the TensorFlow Keras model using the inputs and outputs to your model
model = tf.keras.Model(inputs = inputs, outputs = bounding_box_output)
### END CODE HERE ###
return model
# + [markdown] id="oNWQP3dn1ftJ"
# <a name='ex-05'></a>
# ### Exercise 5
#
# Define the input layer, define the model, and then compile the model.
# - inputs: define an [Input](https://www.tensorflow.org/api_docs/python/tf/keras/Input) layer
# - Set the `shape` parameter. Check your definition of `feature_extractor` to see the expected dimensions of the input image.
# - model: use the `final_model` function that you just defined to create the model.
# - compile the model: Check the [Model](https://www.tensorflow.org/api_docs/python/tf/keras/Model) documentation for how to compile the model.
# - Set the `optimizer` parameter to Stochastic Gradient Descent using [SGD](https://www.tensorflow.org/api_docs/python/tf/keras/optimizers/SGD)
# - When using SGD, set the `momentum` to 0.9 and keep the default learning rate.
# - Set the loss function of SGD to mean squared error (see the SGD documentation for an example of how to choose mean squared error loss).
# + id="C67ZmsTe1n9m"
'''
def define_and_compile_model():
### YOUR CODE HERE ###
# define the input layer
inputs = None
# create the model
model = None
# compile your model
None
### END CODE HERE ###
return model
'''
def define_and_compile_model():
### YOUR CODE HERE ###
# define the input layer
inputs = tf.keras.Input(shape=(224, 224, 3))
# create the model
model = final_model(inputs)
# compile your model
SGD = tf.keras.optimizers.SGD(learning_rate=0.01, momentum=0.9)
model.compile(loss='mean_squared_error', optimizer=SGD)
### END CODE HERE ###
return model
# + [markdown] id="yPtBf83B1zZ3"
# Run the cell below to define your model and print the model summary.
# + id="56y8UNFQIVwj" colab={"base_uri": "https://localhost:8080/"} outputId="6d821c0c-91a9-4802-f0b2-65d7eef1c111"
# define your model
model = define_and_compile_model()
# print model layers
model.summary()
# + [markdown] id="Rypqtdgm6cUa"
# Your expected model summary:
#
# 
# + [markdown] id="HtVVYVlvKXR7"
# <a name='4'></a>
# ## Train the Model
# + [markdown] id="CuhDh8ao8VyB"
# <a name='4.1'></a>
# ### 4.1 Prepare to Train the Model
#
# You'll fit the model here, but first you'll set some of the parameters that go into fitting the model.
#
# - EPOCHS: You'll train the model for 50 epochs
# - BATCH_SIZE: Set the `BATCH_SIZE` to an appropriate value. You can look at the ungraded labs from this week for some examples.
# - length_of_training_dataset: this is the number of training examples. You can find this value by getting the length of `visualization_training_dataset`.
# - Note: You won't be able to get the length of the object `training_dataset`. (You'll get an error message).
# - length_of_validation_dataset: this is the number of validation examples. You can find this value by getting the length of `visualization_validation_dataset`.
# - Note: You won't be able to get the length of the object `validation_dataset`.
# - steps_per_epoch: This is the number of steps it will take to process all of the training data.
# - If the number of training examples is not evenly divisible by the batch size, there will be one last batch that is not the full batch size.
# - Try to calculate the number steps it would take to train all the full batches plus one more batch containing the remaining training examples. There are a couples ways you can calculate this.
# - You can use regular division `/` and import `math` to use `math.ceil()` [Python math module docs](https://docs.python.org/3/library/math.html)
# - Alternatively, you can use `//` for integer division, `%` to check for a remainder after integer division, and an `if` statement.
#
# - validation_steps: This is the number of steps it will take to process all of the validation data. You can use similar calculations that you did for the step_per_epoch, but for the validation dataset.
# + [markdown] id="vgTGU_j-KXR7"
# <a name='ex-06'></a>
# ### Exercise 6
# + id="KoIY6xQ_KXR7"
'''
# You'll train 50 epochs
EPOCHS = 50
### START CODE HERE ###
# Choose a batch size
BATCH_SIZE = None
# Get the length of the training set
length_of_training_dataset = None
# Get the length of the validation set
length_of_validation_dataset = None
# Get the steps per epoch (may be a few lines of code)
steps_per_epoch = None
# get the validation steps (per epoch) (may be a few lines of code)
validation_steps = length_of_validation_dataset//BATCH_SIZE
if length_of_validation_dataset % BATCH_SIZE > 0:
validation_steps += 1
### END CODE HERE
'''
# You'll train 50 epochs
EPOCHS = 50
### START CODE HERE ###
# Choose a batch size
BATCH_SIZE = 64
# Get the length of the training set
length_of_training_dataset = int(len(visualization_training_dataset))
# Get the length of the validation set
length_of_validation_dataset = int(len(visualization_validation_dataset))
# Get the steps per epoch (may be a few lines of code)
steps_per_epoch = 60000//BATCH_SIZE
# get the validation steps (per epoch) (may be a few lines of code)
validation_steps = length_of_validation_dataset//BATCH_SIZE
if length_of_validation_dataset % BATCH_SIZE > 0:
validation_steps += 1
### END CODE HERE
# + [markdown] id="DWCj8CYtSQpY"
# <a name='4.2'></a>
# ### 4.2 Fit the model to the data
#
#
# Check out the parameters that you can set to fit the [Model](https://www.tensorflow.org/api_docs/python/tf/keras/Model#fit). Please set the following parameters.
# - x: this can be a tuple of both the features and labels, as is the case here when using a tf.Data dataset.
# - Please use the variable returned from `get_training_dataset()`.
# - Note, don't set the `y` parameter when the `x` is already set to both the features and labels.
# - steps_per_epoch: the number of steps to train in order to train on all examples in the training dataset.
# - validation_data: this is a tuple of both the features and labels of the validation set.
# - Please use the variable returned from `get_validation_dataset()`
# - validation_steps: teh number of steps to go through the validation set, batch by batch.
# - epochs: the number of epochs.
#
# If all goes well your model's training will start.
# + [markdown] id="BTL8VVAaKXR7"
# <a name='ex-07'></a>
# ### Exercise 7
# + id="TTwH_P-ZJ_xx" colab={"base_uri": "https://localhost:8080/"} outputId="121e7ad4-9eb0-41d3-93b9-928005aed6b2"
'''
### YOUR CODE HERE ####
# Fit the model, setting the parameters noted in the instructions above.
history = None
### END CODE HERE ###
'''
history = model.fit(get_training_dataset(visualization_training_dataset), steps_per_epoch=steps_per_epoch, epochs=10,
validation_data=get_validation_dataset(visualization_validation_dataset), validation_steps=validation_steps)
# + [markdown] id="-aBzmycIsO8w"
# <a name='5'></a>
# ## 5. Validate the Model
#
# <a name='5-1'></a>
# ### 5.1 Loss
#
# You can now evaluate your trained model's performance by checking its loss value on the validation set.
# + id="WWbkUql5sAok" colab={"base_uri": "https://localhost:8080/"} outputId="516e4842-f586-4a78-9639-c3e816bc103b"
loss = model.evaluate(validation_dataset, steps=validation_steps)
print("Loss: ", loss)
# + [markdown] id="Gjtus2EK0-hm"
# <a name='5-2'></a>
# ### 5.2 Save your Model for Grading
#
# When you have trained your model and are satisfied with your validation loss, please you save your model so that you can upload it to the Coursera classroom for grading.
# + id="6Cvv-GgvE3V4"
# Please save your model
model.save("birds.h5")
# + id="CW2AAdkRsOMP" colab={"base_uri": "https://localhost:8080/", "height": 17} outputId="da60fa73-b758-4d36-85be-5ef91db06ae2"
# And download it using this shortcut or from the "Files" panel to the left
from google.colab import files
files.download("birds.h5")
# + [markdown] id="g7E81sgUsUC4"
# <a name='5-3'></a>
# ### 5.3 Plot Loss Function
#
# You can also plot the loss metrics.
# + id="Cz-b8TxU6EDj" colab={"base_uri": "https://localhost:8080/", "height": 283} outputId="01fad6e1-a638-4d9e-82dd-f2c2feebffb9"
plot_metrics("loss", "Bounding Box Loss", ylim=0.2)
# + [markdown] id="5G7KFVX9sXJt"
# <a name='5-4'></a>
# ### 5.4 Evaluate performance using IoU
#
# You can see how well your model predicts bounding boxes on the validation set by calculating the Intersection-over-union (IoU) score for each image.
#
# - You'll find the IoU calculation implemented for you.
# - Predict on the validation set of images.
# - Apply the `intersection_over_union` on these predicted bounding boxes.
# + id="YFqJxt3_VrCm" colab={"base_uri": "https://localhost:8080/"} outputId="83bba596-a184-4a25-bb07-9c9c6c70d226"
def intersection_over_union(pred_box, true_box):
xmin_pred, ymin_pred, xmax_pred, ymax_pred = np.split(pred_box, 4, axis = 1)
xmin_true, ymin_true, xmax_true, ymax_true = np.split(true_box, 4, axis = 1)
#Calculate coordinates of overlap area between boxes
xmin_overlap = np.maximum(xmin_pred, xmin_true)
xmax_overlap = np.minimum(xmax_pred, xmax_true)
ymin_overlap = np.maximum(ymin_pred, ymin_true)
ymax_overlap = np.minimum(ymax_pred, ymax_true)
#Calculates area of true and predicted boxes
pred_box_area = (xmax_pred - xmin_pred) * (ymax_pred - ymin_pred)
true_box_area = (xmax_true - xmin_true) * (ymax_true - ymin_true)
#Calculates overlap area and union area.
overlap_area = np.maximum((xmax_overlap - xmin_overlap),0) * np.maximum((ymax_overlap - ymin_overlap), 0)
union_area = (pred_box_area + true_box_area) - overlap_area
# Defines a smoothing factor to prevent division by 0
smoothing_factor = 1e-10
#Updates iou score
iou = (overlap_area + smoothing_factor) / (union_area + smoothing_factor)
return iou
#Makes predictions
original_images, normalized_images, normalized_bboxes = dataset_to_numpy_with_original_bboxes_util(visualization_validation_dataset, N=500)
predicted_bboxes = model.predict(normalized_images, batch_size=32)
#Calculates IOU and reports true positives and false positives based on IOU threshold
iou = intersection_over_union(predicted_bboxes, normalized_bboxes)
iou_threshold = 0.5
print("Number of predictions where iou > threshold(%s): %s" % (iou_threshold, (iou >= iou_threshold).sum()))
print("Number of predictions where iou < threshold(%s): %s" % (iou_threshold, (iou < iou_threshold).sum()))
# + [markdown] id="9jFVovcUUVs1"
# <a name='6'></a>
# ## 6. Visualize Predictions
#
# Lastly, you'll plot the predicted and ground truth bounding boxes for a random set of images and visually see how well you did!
#
# + id="bR9Bb4uCwTyw" colab={"base_uri": "https://localhost:8080/", "height": 192} outputId="3eecca45-c944-4ba4-9d14-03cc4a345fc8"
n = 10
indexes = np.random.choice(len(predicted_bboxes), size=n)
iou_to_draw = iou[indexes]
norm_to_draw = original_images[indexes]
display_digits_with_boxes(original_images[indexes], predicted_bboxes[indexes], normalized_bboxes[indexes], iou[indexes], "True and Predicted values", bboxes_normalized=True)
# + [markdown] id="lFgrfCW8KXR8"
# <a name='7'></a>
# # 7 Upload your model for grading
#
# Please return to the Coursera classroom and find the section that allows you to upload your 'birds.h5' model for grading. Good luck!
| 192,512 |
/notebooks/NTM_MemDotDeltaQuadraticLowLR.ipynb | d3519c30018409f5bd34d1a3ec96a44d05af3429 | [] | no_license | biseven/LSTM-Optimizer | https://github.com/biseven/LSTM-Optimizer | 0 | 0 | null | null | null | null | Jupyter Notebook | false | false | .py | 578,470 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # REINFORCE
#
# ---
#
# In this notebook, we will train REINFORCE with OpenAI Gym's Cartpole environment.
# ### 1. Import the Necessary Packages
# +
import gym
gym.logger.set_level(40) # suppress warnings (please remove if gives error)
import numpy as np
from collections import deque
import matplotlib.pyplot as plt
# %matplotlib inline
import torch
torch.manual_seed(0) # set random seed
import torch.nn as nn
import torch.nn.functional as F
import torch.optim as optim
from torch.distributions import Categorical
# -
import pdb
# ### 2. Define the Architecture of the Policy
# +
env = gym.make('CartPole-v0')
env.seed(0)
print('observation space:', env.observation_space)
print('action space:', env.action_space)
device = torch.device("cuda:0" if torch.cuda.is_available() else "cpu")
class Policy(nn.Module):
def __init__(self, s_size=4, h_size=16, a_size=2):
super(Policy, self).__init__()
self.fc1 = nn.Linear(s_size, h_size)
self.fc2 = nn.Linear(h_size, a_size)
def forward(self, x):
x = F.relu(self.fc1(x))
x = self.fc2(x)
return F.softmax(x, dim=1)
def act(self, state):
state = torch.from_numpy(state).float().unsqueeze(0).to(device)
probs = self.forward(state).cpu()
m = Categorical(probs)
action = m.sample()
# pdb.set_trace()
return action.item(), m.log_prob(action)
# -
# ### 3. Train the Agent with REINFORCE
policy = Policy().to(device)
optimizer = optim.Adam(policy.parameters(), lr=1e-2)
policy.fc2.weight
def reinforce(n_episodes=1000, max_t=1000, gamma=1.0, print_every=100):
scores_deque = deque(maxlen=100)
scores = []
for i_episode in range(1, n_episodes+1):
saved_log_probs = []
rewards = []
state = env.reset()
for t in range(max_t):
action, log_prob = policy.act(state)
saved_log_probs.append(log_prob)
state, reward, done, _ = env.step(action)
rewards.append(reward)
if done:
break
scores_deque.append(sum(rewards))
scores.append(sum(rewards))
discounts = [gamma**i for i in range(len(rewards)+1)]
R = sum([a*b for a,b in zip(discounts, rewards)])
policy_loss = []
for log_prob in saved_log_probs:
policy_loss.append(-log_prob * R)
policy_loss = torch.cat(policy_loss).sum()
# pdb.set_trace()
optimizer.zero_grad()
policy_loss.backward()
optimizer.step()
if i_episode % print_every == 0:
print('Episode {}\tAverage Score: {:.2f}'.format(i_episode, np.mean(scores_deque)))
if np.mean(scores_deque)>=195.0:
print('Environment solved in {:d} episodes!\tAverage Score: {:.2f}'.format(i_episode-100, np.mean(scores_deque)))
break
return scores
scores = reinforce(n_episodes=400)
# ### 4. Plot the Scores
fig = plt.figure()
ax = fig.add_subplot(111)
plt.plot(np.arange(1, len(scores)+1), scores)
plt.ylabel('Score')
plt.xlabel('Episode #')
plt.show()
# ### 5. Watch a Smart Agent!
# +
env = gym.make('CartPole-v0')
state = env.reset()
for t in range(1000):
action, _ = policy.act(state)
env.render()
state, reward, done, _ = env.step(action)
if done:
break
env.close()
# -
start_lr=0.005)
print(key)
sampler = sample_point_and_params
optimizer_losses[key] = []
for n_iter in [20, 40, 60, 80, 100]:
training_options['n_iter'] = n_iter
# opt.lr.set_value(0.005)
optimizer_losses[key] += opt.train(sampler, **training_options)
# display.clear_output(wait=True)
# opt_loss = np.array(optimizer_loss)
# opt_loss = opt_loss[opt_loss < 10000]
# plt.plot(optimizer_loss)
# -
def get_moving_loss(loss):
moving_loss = [loss[0]]
for i in loss[1:]:
moving_loss.append(0.9 * moving_loss[-1] + 0.1 * i)
return moving_loss
opt_loss = np.array(optimizer_loss)
opt_loss = opt_loss[opt_loss < 10000] / (20. * np.arange(25000)[opt_loss < 10000] / 5000 + 1).astype(int)
plt.plot(opt_loss)
print(opt_loss.max())
opt_loss = np.array(optimizer_loss)
for i in range(5):
cur = opt_loss[i * 5000: (i + 1) * 5000] / (20 * (i + 1))
cur = cur[cur < 10000]
cur = get_moving_loss(cur)
print(cur[-1])
plt.plot(cur, label='{} iterations'.format(20 * (i + 1)))
plt.legend()
non_lstm_optimizers = {
'momentum': momentum_fn
}
# +
# for name, opt in lstm_optimizers.items():
# with open('quadratic_optimizer_drop_coord_low_dim({}).npz'.format(name), 'wb') as f:
# np.savez(f, L.layers.get_all_param_values(opt.l_optim))
# +
# for name, opt in lstm_optimizers.items():
# with np.load('quadratic_optimizer_drop_coord_low_dim({}).npz'.format(name)) as f:
# param_values = [f['arr_%d' % i] for i in range(len(f.files))]
# L.layers.set_all_param_values(opt.l_optim, param_values[0])
# +
import itertools
generalization_loss_lstm = {}
for name in lstm_optimizers:
generalization_loss_lstm[name] = []
# generalization_loss_lstm[name + "_det"] = []
for n_c in itertools.chain(range(2, 150), range(900, 1000)):
print(n_c)
points_and_params = [sample_point_and_params(ndim=n_c) for _ in range(10)]
for name, opt in lstm_optimizers.items():
losses = []
# losses_det = []
for theta, (W_, b_) in points_and_params:
loss = opt.optimize(theta, [W_, b_], 100)[1][-1]
# loss_det = opt.loss_det_fn(theta, 100, W_, b_)[1][-1]
losses.append(loss)
# losses_det.append(loss_det)
generalization_loss_lstm[name].append(np.mean(losses))
# generalization_loss_lstm[name + "_det"].append(np.mean(losses_det))
# +
fig, (ax1, ax2, ax3) = plt.subplots(3, 1, figsize=(12, 24))
for name, losses in sorted(generalization_loss_lstm.items()):
if name.find('False') == -1:
linestyle = 'solid'
else:
linestyle = '--'
ax1.semilogy(list(range(2, 150)) + list(range(900, 1000)), losses[:], label=name, linestyle=linestyle)
ax2.semilogy(list(range(2, 150)), losses[:148], label=name, linestyle=linestyle)
ax3.semilogy(list(range(22, 150)) + list(range(900, 1000)), losses[20:], label=name, linestyle=linestyle)
# fig.legend(loc=4);
ax1.legend(loc=4)
ax2.legend(loc=4)
ax3.legend(loc=2)
fig.tight_layout()
# -
def test_optimizers(**testing_options):
thetas_and_params = [testing_options['sampler']() for _ in range(testing_options['n_functions'])]
histories = {}
for key, opt in lstm_optimizers.items():
print("Testing lstm; {key}".format(**locals()))
loss_history = []
# loss_history_det = []
for theta, (W_, b_) in thetas_and_params:
loss_history.append(opt.optimize(theta, [W_, b_], testing_options['n_iter'])[1])
# loss_history_det.append(opt.loss_det_fn(theta, testing_options['n_iter'], W_, b_)[1])
histories['lstm; {}'.format(key)] = np.median(loss_history, axis=0)
# histories['lstm_det; {}'.format(key)] = np.median(loss_history_det, axis=0)
lrates = np.logspace(0, 29, num=30, base=2.0) * 1e-6
for name, opt in non_lstm_optimizers.items():
best_lrate = None
best_loss = None
best_history = None
print("Testing {name}".format(**locals()))
for lrate in lrates:
loss_history = []
for theta, (W_, b_) in thetas_and_params:
loss_history.append(opt(theta, testing_options['n_iter'], W_, b_, lrate)[1])
if np.isnan(loss_history).any():
break
loss = np.median(loss_history, axis=0)[-1]
if best_loss is None or best_loss > loss:
best_loss = loss
best_lrate = lrate
best_history = np.median(loss_history, axis=0)
histories["{name}; lr={best_lrate}".format(**locals())] = best_history
return histories
# +
testing_options = {
'n_iter': 60,
'n_functions': 50,
'sampler': sample_point_and_params
}
histories_60 = test_optimizers(**testing_options)
# +
testing_options = {
'n_iter': 500,
'n_functions': 50,
'sampler': sample_point_and_params
}
histories_500 = test_optimizers(**testing_options)
# +
testing_options = {
'n_iter': 500,
'n_functions': 1,
'sampler': sample_point_and_params
}
histories_1_func = test_optimizers(**testing_options)
# +
fig, (ax1, ax2, ax3) = plt.subplots(3, 1, figsize=(12, 18))
for name in sorted(set(list(histories_60.keys()) + list(histories_500.keys()) + list(histories_1_func.keys()))):
if name.find('det') != -1:
continue
if name.find('False') == -1:
linestyle = 'solid'
else:
linestyle = '--'
if histories_60.get(name) is not None:
ax1.semilogy(histories_60[name], label=name, linestyle=linestyle)
if histories_500.get(name) is not None:
ax2.semilogy(histories_500[name], label=name, linestyle=linestyle)
if histories_1_func.get(name) is not None:
ax3.semilogy(histories_1_func[name], label=name, linestyle=linestyle)
# fig.legend(loc=4);
ax1.set_title('50 functions; 60 iterations')
ax2.set_title('50 functions; 500 iterations')
ax3.set_title('1 function; 500 iterations')
ax1.legend(loc=4)
ax2.legend(loc=4)
ax3.legend(loc=2)
fig.tight_layout()
# +
theta, (W_, b_) = sample_point_and_params(ndim=2)
sample_runs = {}
for name, opt in lstm_optimizers.items():
history, losses = opt.optimize(theta, [W_, b_], 100)
history = np.concatenate([theta.reshape(1, -1), history], axis=0)
sample_runs[name] = (history, losses)
# history, losses = opt.loss_det_fn(theta, 100, W_, b_)
# history = np.concatenate([theta.reshape(1, -1), history], axis=0)
# sample_runs[name+'_det'] = (history, losses)
sample_runs['momentum'] = momentum_fn(theta, 100, W_, b_, 0.262144)
# +
theta_opt = np.linalg.pinv(W_).dot(b_)
min_x = min(sample_runs[list(lstm_optimizers.keys())[0]][0].T[0])
max_x = max(sample_runs[list(lstm_optimizers.keys())[0]][0].T[0])
min_y = min(sample_runs[list(lstm_optimizers.keys())[0]][0].T[1])
max_y = max(sample_runs[list(lstm_optimizers.keys())[0]][0].T[1])
delta_x = (max_x - min_x) / 100.
delta_y = (max_y - min_y) / 100.
x = np.arange(2 * min_x - (min_x + max_x) / 2, 2 * max_x - (min_x + max_x) / 2, delta_x)
y = np.arange(2 * min_y - (min_y + max_y) / 2, 2 * max_y - (min_y + max_y) / 2, delta_y)
X, Y = np.meshgrid(x, y)
Z = np.zeros(X.shape)
for i in range(X.shape[0]):
for j in range(X.shape[1]):
z = ((W_.dot(np.array([X[i][j], Y[i][j]])) - b_)**2).sum()
Z[i][j] = z
plt.figure(figsize=(15, 12))
plt.title('Trajectory')
CS = plt.contour(X, Y, Z, levels=[1e-4, 1e-3, 1e-2, 5e-2, 1e-1, 5e-1, 1e-0, 5e-0, 1e1])
plt.clabel(CS, inline=1, fontsize=10)
plt.xlabel('x1')
plt.ylabel('x2')
for name in sample_runs:
history, lss = sample_runs[name]
linestyle = 'solid'
plt.plot(history.T[0], history.T[1], linestyle=linestyle, label=name, marker='x')
# history, _ = sample_runs['momentum']
# plt.plot(np.array(history).T[0], np.array(history).T[1], label='momentum', marker='o', linestyle='--')
print(theta_opt)
plt.plot([theta_opt[0]], [theta_opt[1]], marker='x', color='k')
plt.legend();
# +
plt.figure(figsize=(15, 12))
plt.title('loss/step')
plt.xlabel('step')
plt.ylabel('loss')
for name in sample_runs:
_, losses = sample_runs[name]
linestyle = 'solid'
plt.semilogy(losses, label=name, linestyle=linestyle)
plt.legend();
# -
| 12,030 |
/170822.ipynb | cf20f462ecd50325dc049520c60e24d5acca02c0 | [] | no_license | LorinYu/pythonprojectone | https://github.com/LorinYu/pythonprojectone | 0 | 0 | null | null | null | null | Jupyter Notebook | false | false | .py | 1,103 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Python [default]
# language: python
# name: python3
# ---
# # La boucle `for`
#
# La syntaxe est de la forme:
#
# for i in range(début, fin):
# instruction 1
# instruction 2
# ...
#
# Voici quelques exemples:
for i in range(1, 12):
print(i)
# **ATTENTION** Comme vous le voyez l'indice de fin n'est pas inclus dans la boucle.
for i in range(1, 4):
print("Bonjour")
# Par défaut l'indice de départ et 0, si l'indice de début n'est pas précisé.
for i in range(5):
print(i)
# On peut également choisir le pas de l'incrémentation grâce à la syntaxe:
#
# for i in range(début, fin, pas):
# instruction 1
# instruction 2
# ...
for i in range(1, 12, 2):
print(i)
for i in range(365, 300, -5):
print(i)
# *Remarque:* par défaut chaque instruction print se termine par un retour à la ligne, mais on peut suprimer ce retour en choisissant une fin de print vide avec l'attribut end.
for i in range(1, 32):
print(i, end="")
print(" janvier")
# ## Boucles imbriquées
#
# Il est possible d'éxecuter une boucle à l'intérieur d'une autre boucle, elles sont alors dites imbriquées.
for mois in range(1, 13):
for jour in range(1,31):
print(jour, end="/")
print(mois)
# # La boucle `while`
#
# La syntaxe est de la forme:
#
# while condition:
# instruction 1
# instruction 2
# ...
#
# Contrairement à la boucle `for`, le nombre d'itérations n'est pas fixé à l'avance, mais il est fait dynamiquement:
# avant chaque éxecution du bloc d'instructions de la boucle, on test une condition, si cette condition est vérifiée, on éxecute le bloc d'instructiuons de la boucle, sinon, l'éxecution de la boucle est arrétée.
#
# Souvent il faut **initialiser** une variable d'itération avant de commencer la boucle/
#
# *Exemples:*
n = 0
while n < 10:
print(n)
n = n + 1
# Cette boucle est tout à fait identique à la boucle:
#
# for n in range(10):
# print(n)
#
# Cependant une des deux formes est souvent plus adaptée qu'une autre:
#
# - `for` si on connait les itérations à parcourir à l'avance,
#
# - `while` sinon.
#
# En fait, **la boucle `for` est un cas particulier de la boucle `while`**. On peut toujours remplacer une boucle `for` par une boucle `while`, mais l'inverse n'est pas toujours vrai.
#
# **La boucle `while` est un outil plus puissant que la boucle `for`.**
# +
# calcul du logarithme entier (en base 10) d'un nombre x
# Par exemple prenons x = 100
# log(100) = 2 car 10^2 = 100
x = 100
# boucle avec initialisation
n = 0
while x > 1.0:
x = x / 10.0
n = n + 1
print(n)
# -
# ## La non terminaison
#
# Avec la boucle `while`, il est possible de créer un programme qui ne **s'arrête jamais**.
#
# C'est très utile, et me indispensable si l'on considère par exemple les programmes des serveurs de document `html`, qui doivent envoyer les pages `html` indéfiniment à un nombre très important d'utilisateurs.
#
# Cependant, cela peut rendre le debuguage du programme compliqué surtout si on pensait que notre boucle allait s'arrêter mais qu'en fait notre condition d'arrêt était mal implémentée.
#
# En cas de non terminaison involontaire, il faut forcer l'arrêt de l'éxecution du programme.
#
# - dans un notebook sélectionner `Kernel Interrupt` ou `Kernel Restart`
# - dans un terminal python la combinaison de touches <CTRL><C> ou <CTRL><Z> permet souvent d'appliquer un `KeyboardInterrupt`
#
# Par exemple:
n = 3
while n > 0:
print(n, end="")
| 3,737 |
/Studying Training Models.ipynb | 785f00e403991d1fc111695a45801eac42066afc | [] | no_license | Nereland/diamonds-competition | https://github.com/Nereland/diamonds-competition | 0 | 0 | null | null | null | null | Jupyter Notebook | false | false | .py | 52,498 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
import pandas as pd
import seaborn as sns
import matplotlib as plt
import sklearn
from sklearn.preprocessing import StandardScaler, Normalizer
from sklearn.pipeline import make_pipeline
from sklearn import linear_model
from sklearn.model_selection import train_test_split
diam_no = pd.read_csv("outputs/diam_no_out.csv")
diam = pd.read_csv("outputs/diam.csv")
diamfin = pd.read_csv("outputs/diamall.csv")
predict1 = pd.read_csv("outputs/predict1.csv")
predict3 = pd.read_csv("outputs/predict3.csv")
X = diam[["carat","x","y","z","labcol"]]
y = diam["price"]
diamfin
# Best solutions:
# - Normalized
# - With outliers
# ### Normalized data trial final
#
# +
pipeline = [
StandardScaler(),
Normalizer()
]
tr = make_pipeline(*pipeline)
# -
X_readyf = tr.fit_transform(Xf)
X_readyf = pd.DataFrame(X_readyf, columns=Xf.columns)
X_trainf, X_testf, y_trainf, y_testf = train_test_split(X_readyf, yf, test_size=0.2)
# ### Normalized data trial
# +
pipeline = [
StandardScaler(),
Normalizer()
]
tr = make_pipeline(*pipeline)
# -
X_ready = tr.fit_transform(X)
X_ready = pd.DataFrame(X_ready, columns=X.columns)
X_ready.shape
X_ready
X_train, X_test, y_train, y_test = train_test_split(X_ready, y, test_size=0.2)
# ## Linear Regression
diam_model = sklearn.linear_model.LinearRegression()
diam_model.fit(X_train, y_train)
diam_model.score(X_test, y_test)
diam_model_pred = diam_model.predict(X_test)
print("Coefficients: \n", diam_model.coef_)
print("Intercept:", diam_model.intercept_)
y_pred = diam_model.predict(predict1)
y_pred
pred = pd.DataFrame(y_pred)
pred.rename(columns={0: 'price'}, inplace=True)
pred.index.names = ['id']
pred.to_csv("outputs/linearRegression.csv")
# ## RandomForest Regression Model: Descartado
from sklearn.ensemble import RandomForestRegressor
from sklearn.datasets import make_regression
X, y = make_regression(n_features=4, n_informative=2, random_state=0, shuffle=False)
regr = RandomForestRegressor(max_depth=2, random_state=0)
regr.fit(X_train, y_train)
y_predRF = regr.predict(predict1)
#Muy mala puntuación, retocar parámetros
predRF = pd.DataFrame(y_predRF)
predRF.rename(columns={0: 'price'}, inplace=True)
predRF.index.names = ['id']
predRF.to_csv("outputs/RandomForest.csv")
# ## Nearest Neighbors Regression: KNeighborsRegression, interesante
from sklearn.neighbors import KNeighborsRegressor
neigh = KNeighborsRegressor(n_neighbors=2)
neigh.fit(X_train, y_train)
Y_predKNR = neigh.predict(predict1)
predKNR = pd.DataFrame(Y_predKNR)
predKNR.rename(columns={0: 'price'}, inplace=True)
predKNR.index.names = ['id']
predKNR.to_csv("outputs/KNeighborsNormalizedwithout.csv")
# ## Neural Network Regression: interesante
from sklearn.neural_network import MLPRegressor
regr = MLPRegressor(random_state=1, max_iter=500).fit(X_train, y_train)
y_predNN = regr.predict(predict1)
predNN = pd.DataFrame(y_predNN)
predNN.rename(columns={0: 'price'}, inplace=True)
predNN.index.names = ['id']
predNN.to_csv("outputs/NeuralNtwkNORM&OUT.csv")
import h2o
h2o.init(ip="127.0.0.1",max_mem_size_GB = 2)
h2o.init()
# ## Decission Tree Regressor: descartado
from sklearn import tree
clf = tree.DecisionTreeRegressor()
clf = clf.fit(X_train, y_train)
y_predDT = clf.predict(predict1)
predDT = pd.DataFrame(y_predDT)
predDT.rename(columns={0: 'price'}, inplace=True)
predDT.index.names = ['id']
predDT.to_csv("outputs/DecissionTree.csv")
| 3,690 |
/Training a neural network model.ipynb | 300aa9b9ed8c4093c696620bd10bb49828557e92 | [] | no_license | qingwh/Natural-language-processing-and-Text-Mining-with-Python | https://github.com/qingwh/Natural-language-processing-and-Text-Mining-with-Python | 1 | 0 | null | null | null | null | Jupyter Notebook | false | false | .py | 8,402 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# +
import spacy
from spacy.tokens import Doc, Span,Token
from spacy.matcher import Matcher
nlp = spacy.load('en_core_web_sm')
matcher = Matcher(nlp.vocab)
# Two tokens whose lowercase forms match 'iphone' and 'x'
pattern1 = [{'LOWER': 'iphone'}, {'LOWER': 'x'}]
# Token whose lowercase form matches 'iphone' and an optional digit
pattern2 = [{'LOWER': 'iphone'}, {'IS_DIGIT': True, 'OP': '?'}]
# Add patterns to the matcher
matcher.add('GADGET', None, pattern1, pattern2)
# +
TEXTS=['How to preorder the iPhone X', 'iPhone X is coming', 'Should I pay $1,000 for the iPhone X?', 'The iPhone 8 reviews are here', 'Your iPhone goes up to 11 today', 'I need a new phone! Any tips?']
# Create a Doc object for each text in TEXTS
for doc in nlp.pipe(TEXTS):
# Find the matches in the doc
matches = matcher(doc)
# Get a list of (start, end, label) tuples of matches in the text
entities = [(start, end, 'GADGET') for match_id, start, end in matches]
print(doc.text, entities)
# +
TRAINING_DATA = []
# Create a Doc object for each text in TEXTS
for doc in nlp.pipe(TEXTS):
# Match on the doc and create a list of matched spans
spans = [doc[start:end] for match_id, start, end in matcher(doc)]
# Get (start character, end character, label) tuples of matches
entities = [(span.start_char, span.end_char, 'GADGET') for span in spans]
# Format the matches as a (doc.text, entities) tuple
training_example = (doc.text, {'entities': entities})
# Append the example to the training data
TRAINING_DATA.append(training_example)
print(*TRAINING_DATA, sep='\n')
# +
# Create a blank 'en' model
nlp = spacy.blank('en')
# Create a new entity recognizer and add it to the pipeline
ner = nlp.create_pipe('ner')
nlp.add_pipe(ner)
# Add the label 'GADGET' to the entity recognizer
ner.add_label('GADGET')
# -
# Start the training
nlp.begin_training()
import random
# Loop for 10 iterations
for itn in range(10):
# Shuffle the training data
random.shuffle(TRAINING_DATA)
losses = {}
# Batch the examples and iterate over them
for batch in spacy.util.minibatch(TRAINING_DATA, size=2):
texts = [text for text, entities in batch]
annotations = [entities for text, entities in batch]
# Update the model
nlp.update(texts, annotations, losses=losses)
print(losses)
TEST_DATA=['Apple is slowing down the iPhone 8 and iPhone X - how to stop it', "I finally understand what the iPhone X 'notch' is for", 'Everything you need to know about the Samsung Galaxy S9', 'Looking to compare iPad models? Here’s how the 2018 lineup stacks up', 'The iPhone 8 and iPhone 8 Plus are smartphones designed, developed, and marketed by Apple', 'what is the cheapest ipad, especially ipad pro???', 'Samsung Galaxy is a series of mobile computing devices designed, manufactured and marketed by Samsung Electronics']
# Process each text in TEST_DATA
for doc in nlp.pipe(TEST_DATA):
# Print the document text and entitites
print(doc.text)
print(doc.ents, '\n\n')
| 3,349 |
/notpresentation/bagsProcessing.ipynb | ede21b383af539875a184bc436a96310fa9414e3 | [] | no_license | gbarrene/social_tracking | https://github.com/gbarrene/social_tracking | 0 | 0 | null | null | null | null | Jupyter Notebook | false | false | .py | 4,972 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Python 3
# name: python3
# ---
# + [markdown] id="view-in-github" colab_type="text"
# <a href="https://colab.research.google.com/github/dharma610/Project/blob/master/part5_timeseries_plot.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# + id="9geIYv3d-3Rb" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 336} outputId="863bc75c-e257-4090-b397-6dd354e1a656"
from scipy.integrate import odeint
import numpy as np
import pandas as pd
from random import sample
import matplotlib.pyplot as plt
# %matplotlib inline
# !pip install mpld3
import mpld3
mpld3.enable_notebook()
# + id="vGyZypLL-4TT" colab_type="code" colab={}
def deriv(y, t, N, beta, gamma):
S, I, R = y
dSdt = -beta * S * I / N
dIdt = beta * S * I / N - gamma * I
dRdt = gamma * I
return dSdt, dIdt, dRdt
# + id="MhQYzV1y-9VL" colab_type="code" colab={}
def plotsir(t, S, I, R):
f, ax = plt.subplots(1,1,figsize=(14,8))
ax.plot(t, S, 'b', alpha=0.7, linewidth=4, label='Susceptible')
ax.plot(t, I, 'r', alpha=0.7, linewidth=4, label='Infected')
ax.plot(t, R, 'g', alpha=0.7, linewidth=4, label='Recovered')
ax.set_xlabel('Time (days)')
ax.yaxis.set_tick_params(length=0)
ax.xaxis.set_tick_params(length=0)
ax.grid(b=True, which='major', c='w', lw=2, ls='-')
legend = ax.legend()
legend.get_frame().set_alpha(0.5)
for spine in ('top', 'right', 'bottom', 'left'):
ax.spines[spine].set_visible(False)
plt.show();
# + id="T0zByu9t_AxY" colab_type="code" colab={}
""" gamma is a list of 1000 random number in (0,1) """
import random
gamma = []
for i in range(1000):
gamma.append(random.uniform(0, 1))
# + id="SQai82rJBYEj" colab_type="code" colab={}
""" beta is twice of each of 1000 gamma values """
beta = []
for i in range(1000):
beta.append(2*gamma[i])
# + id="cq0qRsEj_DMb" colab_type="code" colab={}
""" P, Q, M are the list of S, I, R respectievely calculated for 100 days for each of 1000 sets of parameter (beta,gamma)."""
P = []
Q = []
M = []
for i in range(1000): #Plotted for 2 sets
N = 1000
S0, I0, R0 = 999, 1, 0 # initial conditions: one infected, rest susceptible
t = np.linspace(0, 99, 100) # Grid of time points (in days)
y0 = S0, I0, R0 # Initial conditions vector
# Integrate the SIR equations over the time grid, t.
ret = odeint(deriv, y0, t, args=(N, beta[i], gamma[i]))
S, I, R = ret.T
P.append(S)
Q.append(I)
M.append(R)
# + id="B3TshUSGPk4k" colab_type="code" colab={}
""" list to dataftrame """
S_value = pd.DataFrame(P)
I_value = pd.DataFrame(Q)
R_value = pd.DataFrame(M)
# + id="dZMw1tL3hKC7" colab_type="code" colab={}
""" S_list, I_list, R_list are the list of S, I, R respectievely chosen for 10 random values of days for each of the rows and there are such 1000 rows,
hence, S_list, I_list, R_list is a list of 1000 itens and each item itself list of 10 values
and S_actual, I_actual, R_actual are list of S, I, R respectively for (day +1) value for each of 10 days for each of 1000 rows """
S_list = []
S_actual = []
I_list = []
I_actual = []
R_list = []
R_actual = []
for i in range(1000):
p = sample(range(0, 99), 10)
S_list.append(S_value[p].iloc[i].to_list())
S_actual.append(S_value[[x+1 for x in p]].iloc[i].to_list())
I_list.append(I_value[p].iloc[i].to_list())
I_actual.append(I_value[[x+1 for x in p]].iloc[i].to_list())
R_list.append(R_value[p].iloc[i].to_list())
R_actual.append(R_value[[x+1 for x in p]].iloc[i].to_list())
# + id="HEoE5Geh9UcJ" colab_type="code" colab={}
""" merging 1000 lists each of 10 items into a single list of 10000 item """
sus_list = []
for i in range(1000):
sus_list = sus_list+S_list[i]
infected_list = []
for i in range(1000):
infected_list = infected_list+I_list[i]
recov_list = []
for i in range(1000):
recov_list = recov_list+R_list[i]
susactual_list = []
for i in range(1000):
susactual_list = susactual_list+S_actual[i]
infectedactual_list = []
for i in range(1000):
infectedactual_list = infectedactual_list+I_actual[i]
recovactual_list = []
for i in range(1000):
recovactual_list = recovactual_list+R_actual[i]
# + id="ktjoaT1cl8jC" colab_type="code" colab={}
""" list to dataframe """
df_1 = pd.DataFrame(sus_list)
df_2 = pd.DataFrame(infected_list)
df_3 = pd.DataFrame(recov_list)
df_4 = pd.DataFrame(susactual_list)
df_5 = pd.DataFrame(infectedactual_list)
df_6 = pd.DataFrame(recovactual_list)
# + id="R53Raf7wpBwz" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 195} outputId="5605c75b-833a-4cb2-f94d-9e390919af05"
""" merging all dataframes into a single dataframe """
data = pd.concat([df_1, df_2,df_3,df_4,df_5,df_6], axis=1)
data.columns = range(data.shape[1])
#data[[1,4]] = data[[1,4]].apply(lambda x: x*10000)
data.head()
# + id="9F860M9RGOxi" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 195} outputId="daac005c-8abf-488a-c13c-49f8595b198e"
from sklearn.preprocessing import MinMaxScaler
scaler = MinMaxScaler()
data_scaled = scaler.fit_transform(data)
unscaled_data = scaler.inverse_transform(data_scaled)
data_scaled = pd.DataFrame(data_scaled)
data_scaled.head()
# + id="3XjM-eOMVo9a" colab_type="code" colab={}
X = data_scaled[[0,1,2]]
y = data_scaled[[3,4,5]]
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=42)
# + id="mJyfSmTonaSx" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 1000} outputId="15e37661-c554-4fda-cf6b-17dc2e5d9652"
from tensorflow import keras
from keras.models import Sequential
from keras.layers import Dense,Activation
from keras.optimizers import Adam
from keras.callbacks import ModelCheckpoint
# load the dataset
# define the keras model
model = Sequential()
model.add(Dense(64, input_dim=3, activation='relu'))
model.add(Dense(32, activation='relu'))
model.add(Dense(32, activation='relu'))
model.add(Dense(32, activation='relu'))
model.add(Dense(32, activation='relu'))
model.add(Dense(32, activation='relu'))
model.add(Dense(16, activation='relu'))
model.add(Dense(16, activation='relu'))
model.add(Dense(3, activation='relu'))
# compile the keras model
opt = keras.optimizers.Adam(learning_rate=0.01)
model.compile(loss='mean_squared_error', optimizer=opt, metrics=['accuracy'])
# checkpoint
filepath="weights-improvement-{epoch:02d}-{val_accuracy:.2f}.hdf5"
checkpoint = ModelCheckpoint(filepath, monitor='val_accuracy', verbose=1, save_best_only=True, mode='max')
callbacks_list = [checkpoint]
# fit the keras model on the dataset
history = model.fit(X_train, y_train, validation_split=0.2, epochs=150, batch_size=10, callbacks=callbacks_list, verbose=0)
# + id="ZpQgexg4L5EQ" colab_type="code" colab={}
predicted_y = model.predict(X_test)
# + id="xmyqlOmkL8Rl" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 34} outputId="09d67ebc-652f-41de-83eb-6986147557ae"
from sklearn.metrics import mean_squared_error
mean_squared_error(y_test, predicted_y)
# + id="_Bap-gouWU-z" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 50} outputId="27795305-79ad-4860-8a5a-5fa6a2f10637"
# evaluate the keras model
_, accuracy = model.evaluate(X_test, y_test)
print('Accuracy: %.2f' % (accuracy*100))
# + [markdown] id="aY8kfgsGFmaV" colab_type="text"
#
# + id="vXhfpbRc0T4N" colab_type="code" colab={}
y_pred1 = model.predict(X_train)
y_pred2 = model.predict(X_test)
df_7 = pd.DataFrame(y_pred1)
df_8 = pd.DataFrame(y_pred2)
# + id="3vgl-yETVq8H" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 195} outputId="27e3af7f-2d00-4b6e-87e7-4788fc236b0b"
y_test.head()
# + id="xkODMVjlK43q" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 326} outputId="3d75125b-0074-49b3-90d7-7879dd1b6af2"
#plot of S_pred1 vs s_actual1
#pred1 corresponds to prediction on training sets , where as actual1 corresponds actual value(y_train)
plt.figure(figsize=(6,4))
plt.plot(df_7[0],y_train[3])
# + id="9sXjGl3c5rqd" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 326} outputId="e02b4c36-feb4-406b-b47b-da117c6dc817"
#plot of I_pred1 vs I_actual1
plt.figure(figsize=(6,4))
plt.plot(df_7[1],y_train[4])
# + id="b7PBM3jRFfP2" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 326} outputId="9ee0e701-0a9b-4326-d19d-c15f9f069355"
plt.figure(figsize=(6,4))
plt.plot(df_7[2],y_train[5]) #plot of R_pred1 vs R_actual1
# + id="U45psDv8Fm9a" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 326} outputId="c2c30943-d16e-40e9-9d4b-78633cbf468b"
#plot of S_pred2 vs S_actual2
#pred2 corresponds to prediction on test sets , where as actual2 corresponds actual value(y_test)
plt.figure(figsize=(6,4))
plt.plot(df_8[0],y_test[3])
# + id="yEToMh95FrN5" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 326} outputId="f6bc2af4-8c12-44fd-f180-99cf10a5d899"
plt.figure(figsize=(6,4))
plt.plot(df_8[1],y_test[4]) #plot of I_pred2 vs I_actual2
# + id="mPfQjGZyFr7w" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 326} outputId="f53f6933-0b20-45be-a5ee-d832a8e97d1d"
plt.figure(figsize=(6,4))
plt.plot(df_8[2],y_test[5]) #plot of R_pred2 vs R_actual2
# + id="DxfCGSSPaoFp" colab_type="code" colab={}
comp_data = pd.DataFrame()
# + id="bAykQT2OuvoS" colab_type="code" colab={}
k = 0
for i in range(100):
comp_data[k] = S_value[i]
comp_data[k+1] = I_value[i]
comp_data[k+2] = R_value[i]
k = k+3
# + id="C5e4FTbMwWXi" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 278} outputId="419581c0-24a5-48a7-c9a9-5a9606186c84"
#original data obtained from solving differential eqn
#columns are in S, I, R order for day 2,3, ......100.
comp_data = comp_data.iloc[:, 3:300]
comp_data.head()
# + id="y68-v1H0xeqw" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 328} outputId="137b83e8-9ded-4967-d507-f0e6c5e1d665"
from sklearn.preprocessing import MinMaxScaler
scaler = MinMaxScaler()
data_scaled_comp = scaler.fit_transform(comp_data)
unscaled_data_comp = scaler.inverse_transform(data_scaled_comp)
data_scaled_comp = pd.DataFrame(data_scaled_comp)
unscaled_data_comp = pd.DataFrame(unscaled_data_comp)
data_scaled_comp.head()
# + id="y1EWDR8NR7Ja" colab_type="code" colab={}
pred_data = {}
pred_data["group0"] = data_scaled_comp[[0,1,2]]
# + id="eKjP-PUQwdLf" colab_type="code" colab={}
# Predicted data
for i in range(98):
pred_data["group" + str(i+1)] = pd.DataFrame(model.predict(pred_data["group" + str(i)]))
# + id="-23q_45k0eVa" colab_type="code" colab={}
predicted_data = pd.DataFrame()
for i in range(98):
predicted_data = pd.concat([predicted_data,pred_data["group" + str(i+1)]],axis = 1)
# + id="Y6Qf44uA5j-q" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 244} outputId="987de50a-d3bc-4279-aad6-0979d8c596dc"
predicted_data.head()
# + id="6BGrIMOLSr0d" colab_type="code" colab={}
original_data = data_scaled_comp.iloc[:,3:297]
# + id="xusEoQBdS9n5" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 328} outputId="19bf6dcd-8838-4f33-e911-28c1ad7915d8"
"""we have to compare between original data and predicted data,
which are data from day 3 to day 100 in S,I,R order"""
original_data.head()
# + id="_ATFl_-ernH5" colab_type="code" colab={}
orig_S = []
for i in range(1,99):
orig_S.append(original_data.iloc[0][3*i])
orig_I = []
for i in range(98):
orig_I.append(original_data.iloc[0][3*i + 4])
orig_R = []
for i in range(98):
orig_R.append(original_data.iloc[0][3*i + 5])
# + id="sb8jYY-io1Mk" colab_type="code" colab={}
t = np.linspace(0, 97, 98) # Grid of time points (in days)
# + id="f1PZZrXKpAAy" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 597} outputId="0ff0cb17-3f17-4593-8217-9a6ada4be96e"
plotsir(t,predicted_data[0].iloc[0] , predicted_data[1].iloc[0], predicted_data[2].iloc[0])
# + id="FUmBhSiHpr4r" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 597} outputId="dcb07ca4-940a-4e51-e3ac-88ed0b2a3ec2"
plotsir(t,orig_S , orig_I, orig_R)
# + id="0v7UDumhtqpH" colab_type="code" colab={}
| 12,609 |
/basic_python/Preliminaries/Chapter2/CONDITIONALS.ipynb | 7a777274a6f4142a938956bdfbc2b59444d5fc69 | [] | no_license | Nhan121/Lectures_notes-teaching-in-VN- | https://github.com/Nhan121/Lectures_notes-teaching-in-VN- | 17 | 5 | null | 2020-04-13T16:07:20 | 2020-04-13T16:06:30 | Jupyter Notebook | Jupyter Notebook | false | false | .py | 21,128 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
from symbulate import *
# %matplotlib inline
# +
states = [1,2]
transition_matrix = [[0.1, 0.9],
[0.7, 0.3]]
initial_dist = [0.5, 0.5]
P = MarkovChainProbabilitySpace(transition_matrix,
initial_dist,
state_labels=states)
X = RV(P)
# -
X.sim(10)
X.sim(1).plot()
X.sim(10).plot()
X[0].sim(10000).plot()
X[1].sim(10000).plot()
X[2].sim(10000).plot()
(X[1] & X[2]).sim(10000).plot(['tile', 'marginal'])
(X[1] & X[3]).sim(10000).plot(['tile', 'marginal'])
(X[3] | (X[1] == 1) ).sim(10000).plot(jitter=True)
X[3].sim(10000).plot()
(X[1] & X[2]).sim(10000).plot(['tile', 'marginal'])
nmax = 4
V = X[range(nmax+1)].apply(count_eq(1)) / (nmax+1)
v = V.sim(10000)
v.plot()
v.mean()
# +
def hitting_time_in_state(sequence, state):
return next( (i for i, x in enumerate(sequence) if x == state), 999999999)
hitting_time_in_state((10,4,5,2,4,5,), 7)
# +
def hitting_time_in_set(sequence, states):
return next( (i for i, x in enumerate(sequence) if x in states), 999999999)
hitting_time_in_set((10,4,5,2,4,5,), [2,5])
# -
T = RV(P, lambda x: hitting_time_in_state(x, 1))
T.sim(10000).tabulate()
the same along all the line of the `conditional block`.
#
# Hence, the `if statement` will raise an `error message` in the following examples:
#
# **Example 1.1.2. No spacing when running `if condition`**
#
# Missing the `TAB` or `spaces` before `your statement` leads to an **`IndentationError`**
a = -3
if a < 0:
print("a is negative")
# So, the **corrrect `if` statement** must be
a = -3
if a < 0:
print("a is negative")
# #### Example 1.1.3. Incorrect number of spaces.
# In this case, the `indentation level` in the third line `print('OK! good job')` has `2 space levels` which is not the same with the previous line: `print("Number %s is a negative"%a)` which has `4 space levels`
if a < 0:
print("Number %s is a negative"%a)
print("OK! good job")
# Again, the **correct one** must be:
if a < 0:
print("Number %s is a negative"%a)
print("OK! good job")
# #### Example 1.1.4. The `statement` is out of `conditional block`
a = -93
if a > 0:
print('%s is positive'%a)
print('Yeah!')
# **Code explaination.**
# - In this case, your `condition` is `a > 0` and the given value `a = -93` which is not satisfied; so, there is no output in the `statement line : print('%s is positive'%a)`!!
# - Moreover, the `final print function` is **out of your `if` statement** so it will be displayed in your output command!
# Now, we know how to write an instruction `block`, we can construct richer conditionals by running some block if the condition is true and an other `block otherwise`. Such a structure is known as a `if ... else statement`.
#
# #### 1.2. The `if-else statement`
#
# Firstly, how to implement the `if-else`'s syntax??
#
# if True_condition:
# Right_statement
# else:
# Wrong_statement
#
# **Example 1.2.1.** The first example on **`if-else`** `statement`!
a = 2018
if a > 0:
print('Condition is true')
print('a is positive')
else:
print('Condition is false')
print('a is nonpositive')
# Quite often, we use a conditional to give a value to a variable according to some condition:
a = 29
if a % 2 == 1:
val = 'Odd number'
else:
val = 'Even number'
print(val)
# Such a code is a bit long and to reduce its size, **`Python`** offers ***inline conditional***:
#
# (Right_statement) if (True_condition) else (Wrong_statement)
val = 29
val = 'Odd number' if (a % 2 == 1) else 'Even number'
print(val)
# An **`if ... else`** `statement` is adapted to test a `single condition` but it becomes ***less easy when multiple nested conditions are needed***.
#
# **Example 1.2.2.** Write an `if-else statement` to print out the results of the `remainder` in the `quotient` by `4`.
#
# We have known that
#
# a % 4 == 0 then remainder = 0
# a % 4 == 1 then remainder = 1
# a % 4 == 2 then remainder = 2
# a % 4 == 3 then remainder = 3
#
# In case of using only **`if-else`** `statement`, this really take a lots of lines in your condition and statement.
#
# if a % 4 == 0
# remainder = 0
# else: ## this cass a % 4 == 1 or == 2 or == 3
# ## then this need to an if-else inside the else's statement
# if a % 4 == 1:
# remainder = 1
# else:
# ## this time; the remainders can be 2 or 3
# if a % 4 == 2:
# remainder = 2
# else:
# remainder = 3
# Look at the following code!
if a % 4 == 0:
res = 0
else:
if a % 4 == 1:
res = 1
else:
if a % 4 == 2:
res = 2
else:
res = 4
print("The remainder of %s by 4 is %s"%(a, res))
# To produce a code easier to read and test the various conditions one by one, we can deal with a **`if ... elif ... else`** statement.
#
# #### 1.3. The `if ... elif ... else` statement
# The **syntax** be
#
# if (your first condition):
# first_statement
# elif (your 2nd condition):
# 2nd_statement
# ...
# elif (your k th condition):
# k_th_statement
# else:
# last_statement
# So, the previous example can elegantly be rewritten as follows:
#
# **Example 1.3.1.** Rewrite the preceding code using `if-elif-else` statement!
if a % 4 == 0:
res = 0
elif a % 4 == 1:
res = 1
elif a % 4 == 2:
res = 2
else:
res = 3
print("The remainder of %s by 4 is %s"%(a, res))
# **Example 1.3.2.** Using `if-elif-else` statement to named the `month` based on the integer inputs. For example:
# - `input = 1` then `output = "January`
# - `input = 10` then `output = October`
# +
month_int = 8
if month_int == 1:
month_name = "January"
elif month_int == 2:
month_name = "February"
elif month_int == 3:
month_name = "March"
elif month_int == 4:
month_name = "April"
elif month_int == 5:
month_name = "May"
elif month_int == 6:
month_name = "June"
elif month_int == 7:
month_name = "July"
elif month_int == 8:
month_name = "August"
elif month_int == 9:
month_name = "September"
elif month_int == 10:
month_name = "October"
elif month_int == 11:
month_name = "November"
else:
month_name = "December"
print("month_int = %s, month_name = %s"%(month_int, month_name))
# -
# But, we see that using `if-elif-else` in this case remained many complexities in your code, isn't it? To make it more simplifier, we will introduce another approach : **`switch-case`** `statement`!
#
# ## 2. Switch-case statement!
#
# **Syntax:**
#
# switcher = {
# case 1: statement_1,
# case 2: statement_2,
# ... .... ,
# case n: statement_n
# }
# switcher.get(args, "default statement (otherwise)")
switcher = {
1: "January",
2: "February",
3: "March",
4: "April",
5: "May",
6: "June",
7: "July",
8: "August",
9: "September",
10: "October",
11: "November",
12: "December"
}
# Now, verify our results with the `input = 8, 10, 14` repestively, and we expect that the `input = 14` returns the `"invalid month"`!
month_int = 8; print(switcher.get(month_int, "invalid month"))
month_int = 10; print(switcher.get(month_int, "invalid month"))
month_int = 14; print(switcher.get(month_int, "invalid month"))
# ## 3. Practices & exercises.
#
# **Exercise 3.1.** Being given some variable `v`, write a conditional to test if `v` is **`None`** or not!
#
# **SOLUTION.**
#
# Firstly, we will verify with `v = 3` or `v` is not **`None`**!
v = 3
if v == None:
print("Exactly!! v is None")
else:
print("Fuck!! v is not None")
# and in case `v` is **`None`**!
v = None
if v == None:
print("Exactly!! v is None")
else:
print("Fuck!! v is not None")
# **Exercise 3.2.** Being given two `strings: s1 and s2`, write a conditional to test if they have the same length or not?
#
# **SOLUTION.**
#
# The behind idea is using the `function` `len()` to find exactly the length of a `string`!
# +
s1 = "Hello!! My name is Nhan and I graduated Master in Paul Sabatier, France!!"
s2 = "I have been a Data Scientist and Machine Learning Engineering for 2 years."
if len(s1) == len(s2):
print("the string s1 is the same length with the s2 string")
else:
print("Not equal! Since length(s1) = %s, while length(s2) = %s"%(len(s1), len(s2)))
# -
# **Exercise 3.3.** For some numeric value `v`, write a **`if ... elif ... else`** `statement` that prints `different messages` according to the `positivity, nullity` or `negativity` of `v`.
v = 2015.5
if v > 0:
print("%s is positivity"%v)
elif v < 0:
print("%s is negativity"%v)
else:
print("%s is nullity"%v)
# **Exercise 3.4.** Using the `if-elif-else` `statement` to define the `numbers of day` in a given `month`, `year`? For example:
#
# | input: `year` | input: `month` | output : `numbers_of_day` |
# |-|-|-|
# | 2001 | 3 | 31|
# | 2001 | 2 | 28 |
# | 2000 | 2 | 29 |
# +
month = 2
year = 2001
if (month == 2) and (year % 4 == 0):
numbers_of_day = 29
elif (month == 2) and (year % 4 != 0):
numbers_of_day = 28
elif month in [4, 6, 9, 11]:
numbers_of_day = 30
else:
numbers_of_day = 31
print("Mon = %s, year = %s has %s days"%(month, year, numbers_of_day))
# -
# Another checking in loop year!
# +
month = 2; year = 2000
if (month == 2) and (year % 4 == 0):
numbers_of_day = 29
elif (month == 2) and (year % 4 != 0):
numbers_of_day = 28
elif month in [4, 6, 9, 11]:
numbers_of_day = 30
else:
numbers_of_day = 31
print("Mon = %s, year = %s has %s days"%(month, year, numbers_of_day))
# -
# Final checking!
# +
month = 3; year = 2001
if (month == 2) and (year % 4 == 0):
numbers_of_day = 29
elif (month == 2) and (year % 4 != 0):
numbers_of_day = 28
elif month in [4, 6, 9, 11]:
numbers_of_day = 30
else:
numbers_of_day = 31
print("Mon = %s, year = %s has %s days"%(month, year, numbers_of_day))
| 10,839 |
/tensorflow/practice_tf.ipynb | 92b85a88224d790be822e0b543b49edab5e1a9a8 | [] | no_license | jakeoung/jkopt | https://github.com/jakeoung/jkopt | 0 | 0 | null | null | null | null | Jupyter Notebook | false | false | .py | 12,925 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Day 2 Assignment
# Q.1) List and its default functions.
lst = ["Tejal",10,7186,126.43,[1,2,3]]
print(lst)
lst[1]
lst[4]
lst[4][0]
lst[4][2]
lst.append("kiran")
print(lst)
lst.index(10)
lst[4]
lst[4][2]
# Q.2) Dictionary and its default functions.
dit = {"name": "Tejal","age": "22", "number": 123456, "email": "[email protected]"}
print(dit)
# # Methods of DICT
dit["name"]
dit.items()
dit.keys()
dit.pop("name")
dit
dit["school"]="KES"
dit
# Q.3) Sets and its default functions.
st = {"Tej","letsupgrade",1,2,3,4,4,5,6,5,5}
st
st1 = {"Tej",1}
st1.issubset(st)
# Q.4) Tuple and explore default methods.
tup = ("Tej","&","Letspgrade.in")
tup
tup.count("&")
tup.index("Letspgrade.in")
# Q.5) String and explore default methods.
a="hello"
print(a)
a[1]
a[3]
# # String Methods:
# Count
str1 = "what we thinl we become"
str1
str1.count("we")
str1.count("we",0,15)
# Find
str1
str1.find("we")
str1.rfind("we")
# String case methods:
# lowercase()
str2 = "TEJAL"
str2.lower()
# uppercase()
a="hello"
a.upper()
# Index
a
a=a.upper()
a
a.index("H")
val = input("Enter the string")
if val in a :
print(a.index(val))
# Capitalize
str1
str1.capitalize()
# title
str1.title()
# swapcase()
str3 = "HELLO"
str3.swapcase()
# Strip
str3 = "##$intel#$"
str3
str3.strip("#$")
str3.rstrip("#$")
str3.lstrip("#$")
# split
date1 = "16-22-2020"
date1[6:]
date1.split("-")
date1.split("-")[-1]
ip = "192.168.10.1"
ip.split(".")
ip.split(".",1)
| 1,783 |
/Handwritten digits recognition.ipynb | e1ac6e2efec2267cced351d8eaebc638fe804984 | [] | no_license | rishitshah12/Handwritten-Digit-recognition-using-CNN-algorithm | https://github.com/rishitshah12/Handwritten-Digit-recognition-using-CNN-algorithm | 0 | 0 | null | null | null | null | Jupyter Notebook | false | false | .py | 60,865 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
from tqdm import tqdm
import tflearn
import tensorflow as tf
import pandas as pd
import numpy as np
import os
import cv2
from tflearn.layers.conv import conv_2d,max_pool_2d
from tflearn.layers.core import input_data,fully_connected,dropout
from tflearn.layers.estimator import regression
import matplotlib.pyplot as plt
from skimage import color
from skimage import io
digits = pd.read_csv('/home/rajjjj/Downloads/mnist_train.csv')
digits_test = pd.read_csv('/home/rajjjj/Downloads/mnist_test.csv')
#labels
ytrain1 = digits['label']
#features
xtrain = digits.drop(['label'],axis=1)
xtrain = xtrain.values
ytrain1 = ytrain1.values
#labels
ytest1 = digits_test['label']
ytest1 = ytest1.values
#features
xtest = digits_test.drop(['label'],axis=1)
xtest = xtest.values
# +
ytest = []
ytrain = []
for i in range(len(ytest1)):
if(ytest1[i]==0):
ytest.append([1,0,0,0,0,0,0,0,0,0])
elif(ytest1[i]==1):
ytest.append([0,1,0,0,0,0,0,0,0,0])
elif(ytest1[i]==2):
ytest.append([0,0,1,0,0,0,0,0,0,0])
elif(ytest1[i]==3):
ytest.append([0,0,0,1,0,0,0,0,0,0])
elif(ytest1[i]==4):
ytest.append([0,0,0,0,1,0,0,0,0,0])
elif(ytest1[i]==5):
ytest.append([0,0,0,0,0,1,0,0,0,0])
elif(ytest1[i]==6):
ytest.append([0,0,0,0,0,0,1,0,0,0])
elif(ytest1[i]==7):
ytest.append([0,0,0,0,0,0,0,1,0,0])
elif(ytest1[i]==8):
ytest.append([0,0,0,0,0,0,0,0,1,0])
elif(ytest1[i]==9):
ytest.append([0,0,0,0,0,0,0,0,0,1])
for i in range(len(ytrain1)):
if(ytrain1[i]==0):
ytrain.append([1,0,0,0,0,0,0,0,0,0])
elif(ytrain1[i]==1):
ytrain.append([0,1,0,0,0,0,0,0,0,0])
elif(ytrain1[i]==2):
ytrain.append([0,0,1,0,0,0,0,0,0,0])
elif(ytrain1[i]==3):
ytrain.append([0,0,0,1,0,0,0,0,0,0])
elif(ytrain1[i]==4):
ytrain.append([0,0,0,0,1,0,0,0,0,0])
elif(ytrain1[i]==5):
ytrain.append([0,0,0,0,0,1,0,0,0,0])
elif(ytrain1[i]==6):
ytrain.append([0,0,0,0,0,0,1,0,0,0])
elif(ytrain1[i]==7):
ytrain.append([0,0,0,0,0,0,0,1,0,0])
elif(ytrain1[i]==8):
ytrain.append([0,0,0,0,0,0,0,0,1,0])
elif(ytrain1[i]==9):
ytrain.append([0,0,0,0,0,0,0,0,0,1])
# -
ytest = np.array(ytest)
ytrain = np.array(ytrain)
# +
xtest1 = xtest.reshape(-1,28,28)
xtrain1 = xtrain.reshape(-1,28,28)
# +
#input layer
cnet=input_data(shape=[None,28,28,1],name='input')
#1st layer
cnet=conv_2d(cnet,26,3,regularizer='L1')
cnet=max_pool_2d(cnet,3)
#2nd layer
cnet=conv_2d(cnet,24,3,regularizer='L1')
cnet=max_pool_2d(cnet,3)
#3rd layer
cnet=conv_2d(cnet,22,3,regularizer='L1')
cnet=max_pool_2d(cnet,3)
#4th layer
cnet=conv_2d(cnet,20,3,regularizer='L1')
cnet=max_pool_2d(cnet,3)
#fully
cnet=fully_connected(cnet,324)
#dropout
cnet=dropout(cnet,keep_prob=0.6)
# output
cnet=fully_connected(cnet,10,activation='softmax')
# -
cnet=regression(cnet,optimizer='adam',loss='categorical_crossentropy',name='output',learning_rate=0.0003)
cnmodel = tflearn.DNN(cnet)
xtest = []
xtrain = []
for i in range(len(xtest1)):
xtest.append(color.rgb2gray(xtest1[i]))
for i in range(len(xtrain1)):
xtrain.append(color.rgb2gray(xtrain1[i]))
xtest = np.array(xtest).reshape(-1,28,28,1)
xtrain = np.array(xtrain).reshape(-1,28,28,1)
# ytest = ytest.reshape(-1,1)
# ytrain = ytrain.reshape(-1,1)
# ytrain.shape
xtest[0]
cnmodel.fit({'input':xtrain.reshape(-1,28,28,1)},{'output':ytrain},
n_epoch=2,
validation_set=({'input':xtest},
{'output':ytest}),
show_metric=True)
cnmodel.save('digits.tf1')
np.argmax(cnmodel.predict(xtest[500].reshape(1,28,28,1)))
np.argmax(ytest[500])
plt.imshow(xtest[500].reshape(28,28),cmap='gray')
import cv2
f = cv2.resize(xtest[500],(200,200))
xtrain[0]
| 4,078 |
/线性回归评价标准.ipynb | c071a757aa726c56226eaeca8173c26a7751355c | [] | no_license | setsuren/vs1 | https://github.com/setsuren/vs1 | 3 | 0 | null | null | null | null | Jupyter Notebook | false | false | .py | 11,708 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Importing Libraries
#
# This might take a few seconds. If any library is missing, make sure to install it in your environment, using anaconda in for library installation is usually easier
# +
import numpy as np
import matplotlib.pyplot as plt
x, y = np.random.random(size=(2,10))
# print(x[0:0+2])
print(x)
print(y)
for i in range(0, len(x),2):
print(x[i:i+2])
print(y[i:i+2])
plt.plot(x[i:i+2], y[i:i+2], 'o-')
plt.show()
# -
import matplotlib
from mpl_toolkits import mplot3d
from matplotlib import pyplot as plt
from matplotlib import cm
from matplotlib import image as mpimg
from matplotlib.pyplot import figure
import seaborn as sns
import numpy as np
import matplotlib.pylab as pl
from matplotlib.colors import ListedColormap
# %matplotlib notebook
# +
import matplotlib.pyplot as plt
import numpy as np
# An "interface" to matplotlib.axes.Axes.hist() method
d = np.random.laplace(loc=15, scale=3, size=500)
d[:5]
n, bins, patches = plt.hist(x=d, bins='auto', color='#0504aa',
alpha=0.7, rwidth=0.85)
plt.grid(axis='y', alpha=0.75)
plt.xlabel('Value')
plt.ylabel('Frequency')
plt.title('My Very Own Histogram')
plt.text(23, 45, r'$\mu=15, b=3$')
maxfreq = n.max()
# Set a clean upper y-axis limit.
plt.ylim(top=np.ceil(maxfreq / 10) * 10 if maxfreq % 10 else maxfreq + 10)
# -
# # Introduction
#
# This section is meant as a basic intro. There are no graded sections in this part
# # 1. Loading an Image
# +
# reading image from file to memory
img = np.array(mpimg.imread('intro/cat.jpg'))
# allows us to manipulate the image
img.setflags(write=1)
# -
# We can see that the <b>img</b> variable is a 3D array. In other words, it is a 2D array of pixels with values for rgb. The variable for each color is an 8 bit integer ranging from 0-255.
img
# # 2. Displaying an Image
# we can display an image just by using the `plt.imshow` function.
#
# we use the <a href="https://matplotlib.org/">matplotlib</a> library to display our image in jupyter notebooks
# %matplotlib notebook
figure(dpi = 150)
plt.axis('off') # allows us to turn off axis, comment this out to show axis
plt.imshow(img)
plt.show()
# # 3 Image Manipulation
# ## 3.1 crop
#
# we can crop an just by displaying a subsection of the array, here i've focused the crop section on the cat's head
# %matplotlib notebook
figure(dpi = 150)
plt.axis('off')
plt.imshow(img[100:600,650:1300])
plt.show()
# ## 3.2 Markers
#
# We can create markers on our image. Here i'm displaying 50 randomly placed markers.
#
# you can access other marker types at https://matplotlib.org/api/_as_gen/matplotlib.pyplot.plot.html
# +
# generate random integer values
import random
from random import seed
from random import randint
# seed random number generator
seed(1)
# %matplotlib notebook
figure(dpi = 150)
plt.axis('off')
plt.imshow(img) #drawing image
for i in range(50):
plt.plot( randint(0,img.shape[1]-1), randint(0,img.shape[0]-1), 'b+') #adding markers to plot
plt.show()
# -
# ## 3.3 Color Analysis
#
#
# #### Plotting in 2d
#
# We can individually access the rgb values and we plot them. We see that blue is considerably left shifted, which is evident from the image which is mostly green or red
#
# We can use the <a href="https://seaborn.pydata.org/examples/index.html">seaborn</a> library for displaying graphs
# +
#referencing individual rgb values
r = img[...,0]
g = img[...,1]
b = img[...,2]
figure(dpi = 150)
#drawing dist plots
ax = sns.distplot(np.reshape(r, -1), hist=True, color = 'r', kde_kws={"shade": False})
ax = sns.distplot(np.reshape(g, -1), hist=True, color = 'g', kde_kws={"shade": False})
ax = sns.distplot(np.reshape(b, -1), hist=True, color = 'b', kde_kws={"shade": False})
#displaying plot
ax.set(xlabel='Color Value', ylabel='M of pixel')
plt.show()
# -
# #### Plotting in 3d
#
# If we want to visualize the color space in 3d, we can do that using `matplotlib`
#
# if youre interested in exploring the 3d properties of `matplotlib`, explore the <a href="https://matplotlib.org/tutorials/toolkits/mplot3d.html#scatter-plots">mplot3d</a> library
#
# you might find this site helpful too: https://jakevdp.github.io/PythonDataScienceHandbook/04.12-three-dimensional-plotting.html
# +
# %matplotlib notebook
fig = figure(dpi = 150)
ax = plt.axes(projection='3d')
# converting the 2d rgb values to 1d arrays
r = np.reshape(img[...,0],-1)
g = np.reshape(img[...,1],-1)
b = np.reshape(img[...,2],-1)
# chosing a random subset of the image data, we do not want to plot the enitre pixel values as it it'll take forever
i_rand = np.random.choice(range(len(r)), 1000, replace= False)
# Data for three-dimensional scattered points
ax.scatter3D(r[i_rand],
g[i_rand],
b[i_rand],
c=r[i_rand],
cmap = "gray")
ax.set(xlabel='r', ylabel='g', zlabel='b')
# -
# ## 3.4 Color Manipulation, Viewing Red
#
# Suppose we want to just view the red values. We can do this by setting the green and blue values to 0.
def rgb2red(rgb):
img_red = rgb.copy() # we make a copy because we DO NOT want to manipulate the original array
img_red[:,:,1:3] = 0 # we chose all rows and columns and set the 2nd and 3rd pixel values to 0
return img_red
img_red = rgb2red(img)
figure(dpi = 150)
plt.axis('off')
plt.imshow(img_red)
plt.show()
# ## 3.5 Color Manipulation, Viewing Gray
#
# Suppose now we want to view the image in grayscale. We can use the previous method of pixel manipulation.
#
# the grey value is given by the formula <a href="https://en.wikipedia.org/wiki/Grayscale#Converting_color_to_grayscale"><b>y = 0.299r + 0.587g + 0.114b</b></a>
#
#
def rgb2gray(rgb):
return np.uint8(np.dot(rgb[...,:3], [0.299, 0.587, 0.114]))
img_gray = rgb2gray(img)
img_gray #here we see the image as a 2D aray of a sigle 8bit integer
figure(dpi = 150)
plt.axis('off')
plt.imshow(img_gray,cmap='gray') # cmap sets the color map to gray
plt.show()
# # 0.4. Saving Image
#
# We can easily save an image by the <b>mpimg.imsave</b> function. The function automatically detects file type from the name
mpimg.imsave('intro/cat_gray.jpg',img_gray,cmap='gray') # if saving rgb files, remove cmap
# # 5. Test Yourself
#
# I want to saturate the image by adding 100 values to the pixels in the image. I make the following function to add 100 to each pixel value
def add_100(rgb):
added_100 = img.copy() + 100
return added_100
added_100 = add_100(img)
# #### Something's wrong! I ran the function but it doesnt display the image properly! what's wrong with my funtion?
figure(dpi = 150)
plt.axis('off')
plt.imshow(added_100)
plt.show()
# ### The function should display the following image
#
# fix the function to display the proper image
figure(figsize=(30, 30))
plt.axis('off')
plt.imshow(added_100)
plt.show()
| 7,092 |
/compas/code/.ipynb_checkpoints/DToolsCOMPAS-checkpoint.ipynb | 3f147efa001011a767486e1e7efa25877b255eb6 | [
"Apache-2.0"
] | permissive | shubhampachori12110095/nips2017-1 | https://github.com/shubhampachori12110095/nips2017-1 | 0 | 0 | null | null | null | null | Jupyter Notebook | false | false | .py | 1,487,780 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
import torch
from torch.utils.data import DataLoader
from torch import nn, optim
from torchvision import transforms, datasets
import visdom
class VAE(nn.Module):
def __init__(self):
super(VAE, self).__init__()
# [b, 784] => [b, 20]
# u: [b, 10]
# sigma: [b, 10]
self.encoder = nn.Sequential(
nn.Linear(784, 256),
nn.ReLU(),
nn.Linear(256, 64),
nn.ReLU(),
nn.Linear(64, 20),
nn.ReLU()
)
# [b, 20] => [b, 784]
self.decoder = nn.Sequential(
nn.Linear(10, 64),
nn.ReLU(),
nn.Linear(64, 256),
nn.ReLU(),
nn.Linear(256, 784),
nn.Sigmoid()
)
self.criteon = nn.MSELoss()
def forward(self, x):
"""
:param x: [b, 1, 28, 28]
:return:
"""
batchsz = x.size(0)
# flatten
x = x.view(batchsz, 784)
# encoder
# [b, 20], including mean and sigma
h_ = self.encoder(x)
# [b, 20] => [b, 10] and [b, 10]
mu, sigma = h_.chunk(2, dim=1)
# reparametrize trick, epison~N(0, 1)
std = torch.exp(sigma)*0.5
h = mu + std * torch.randn_like(sigma)
# decoder
x_hat = self.decoder(h)
# reshape
x_hat = x_hat.view(batchsz, 1, 28, 28)
kld = torch.sum(torch.exp(sigma)+torch.pow(mu, 2) -sigma-1) / (batchsz*28*28)
return x_hat, kld
# +
mnist_train = datasets.MNIST('data', True, transform=transforms.Compose([
transforms.ToTensor()
]), download=True)
mnist_train = DataLoader(mnist_train, batch_size=32, shuffle=True)
mnist_test = datasets.MNIST('data', False, transform=transforms.Compose([
transforms.ToTensor()
]), download=True)
mnist_test = DataLoader(mnist_test, batch_size=32, shuffle=True)
x, _ = iter(mnist_train).next()
print('x:', x.shape)
# device = torch.device('cuda')
# model = VAE().to(device)
model = VAE()
criteon = nn.MSELoss()
optimizer = optim.Adam(model.parameters(), lr=1e-3)
# print(model)
viz = visdom.Visdom()
for epoch in range(1000):
for batchidx, (x, _) in enumerate(mnist_train):
# [b, 1, 28, 28]
# x = x.to(device)
x_hat, kld = model(x)
loss = criteon(x_hat, x)
if kld is not None:
loss = loss+0.5*kld
# backprop
optimizer.zero_grad()
loss.backward()
optimizer.step()
print(epoch, 'loss:', loss.item(), 'kld:', kld.item())
x, _ = iter(mnist_test).next()
# x = x.to(device)
with torch.no_grad():
x_hat, kld = model(x)
viz.images(x, nrow=8, win='x', opts=dict(title='x'))
viz.images(x_hat, nrow=8, win='x_hat', opts=dict(title='x_hat'))
# -
a= 'abc'
a[0:5]
| 3,103 |
/projeto_ml_0610_10h39.ipynb | b9c6914e38a00991b8b10a8de41a662ee28d44e8 | [] | no_license | FernandoHonda1/projeto_ml_draft | https://github.com/FernandoHonda1/projeto_ml_draft | 0 | 0 | null | null | null | null | Jupyter Notebook | false | false | .py | 2,640,909 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# +
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import pandas as pd
from scipy.stats import pearsonr
from scipy.stats import chi2_contingency
from scipy.stats import chi2
from sklearn import tree
from sklearn.tree import DecisionTreeClassifier
from sklearn.impute import SimpleImputer
from sklearn.linear_model import LogisticRegression
from sklearn.metrics import roc_auc_score
df = pd.read_pickle('treino.pkl.gz')
# +
# função 'chamada' por 'proba_box'
def discretize_proba(x, thresh):
for n in range(len(thresh)):
if x == thresh[n]:
return n + 1
# discretiza feature quantitativa, utilizando árvores de decisão, armazena novas categorias em outra feature,
# plota box plot de acordo com taxa de default em cada nova categoria, gerada na discretização
# apesar de criar nova feature no dataframe passado, suponho que não haja problema, por exemplo, rodar
# duas vezes em sequência, com diferentes n(max_depth), uma vez que, as novas features são criadas com base
# na feature de nome 'series_name', que permanece inalterada, o que acontece, caso esta função seja 'chamada'
# diversas vezes é que a nova feature é constantemente redefinida
def proba_box(dataframe, series_name, target, max_depth, random_state):
disc = DecisionTreeClassifier(max_depth = max_depth, random_state = random_state)
disc.fit(dataframe[[series_name]], dataframe[[target]])
dataframe[series_name + '_proba'] = disc.predict_proba(dataframe[[series_name]])[:, 1]
thresh = dataframe[series_name + '_proba'].unique().tolist()
thresh.sort()
dataframe[series_name + '_proba'] = dataframe[series_name + '_proba'].apply(lambda x:
discretize_proba(x, thresh))
plt.style.use('default')
plt.rcParams['figure.figsize'] = (11, 5)
sns.boxplot(dataframe[series_name + '_proba'], dataframe[series_name])
plt.show()
# visualizar árvore de decisão (permite analisar como foi feita a discretização)
def tree_thresholds(dataframe, series_name, target, max_depth, random_state):
mdl = DecisionTreeClassifier(random_state = random_state, max_depth = max_depth)
mdl.fit(dataframe[[series_name]], dataframe[[target]])
plt.style.use('default')
fig = plt.figure(figsize = (12, 5))
_ = tree.plot_tree(mdl)
plt.show()
# roda funções tree_thresholds e proba_box em conjunto, para quantas camadas de árvore forem solicitadas
def tree_box_plot(dataframe, series_name, target, iterations, random_state):
for i in iterations:
tree_thresholds(dataframe, series_name, target, i, random_state)
proba_box(dataframe, series_name, target, i, random_state)
def target_prop_bin(dataframe, series_name, target):
plot_data = []
for i in dataframe[series_name].unique():
data = dataframe.loc[dataframe[series_name] == i, target].value_counts(1)
if len(data) == 2:
plot_data.append([str(i), data.iloc[1]])
if len(data) < 2:
plot_data.append([str(i), data.iloc[0]])
plot_data.sort()
for i in plot_data:
plt.bar(i[0], i[1], color = 'turquoise')
plt.xticks(range(len(plot_data)), [i[0] for i in plot_data])
plt.xticks(rotation = 45)
plt.yticks([])
plt.show()
# teste de independência entre variáveis categóricas
def quali_var_indep_hyp_test(dataframe, series_name, target, alpha):
stat, pval, dof, exp = chi2_contingency(pd.crosstab(dataframe[series_name], dataframe[target]))
crit = chi2.ppf(q = (1 - alpha), df = dof)
if stat >= crit:
cramer = np.sqrt( ( stat ) / sum(sum(exp)) * (min(exp.shape) - 1) )
else:
cramer = None
return stat, crit, cramer
# tranformar feature 'nascimento' em 'idade'
def year_to_age(x):
if x == '00':
return 20
else:
return (100 - int(x)) + 20
# transformar formato e unidade das features 'tem_med_emp' e 'tem_pri_emp'
def time_unit_unification(x):
sub1 = x.split(' ')[0]
sub2 = x.split(' ')[1]
return (int(sub1.strip('yrs')) * 12) + int(sub2.strip('mon'))
plt.style.use('dark_background')
plt.rcParams['figure.figsize'] = (16, 8)
# +
# seleção preliminar de features:
# o critério desta foi: lógica / explicabilidade, as features eliminadas são consideradas como não relacionadas
# à probabilidade de 'deafult' ou não
sub1 = ['id_pessoa', 'valor_emprestimo', 'custo_ativo', 'emprestimo_custo', 'nascimento', 'emprego', 'score',
'score_desc']
sub2 = ['tem_med_emp', 'tem_pri_emp']
sub4 = ['pri_qtd_tot_emp', 'pri_qtd_tot_emp_atv', 'pri_qtd_tot_def', 'pri_emp_abt', 'pri_emp_san','pri_emp_tom']
sub5 = ['sec_qtd_tot_emp', 'sec_qtd_tot_emp_atv', 'sec_qtd_tot_def', 'sec_emp_abt', 'sec_emp_san','sec_emp_tom']
# sub1_dropd = ['agencia', 'revendedora', 'montadora', 'Current_pincode_ID', 'data_contrato', 'estado',
# 'funcionario']
# sub2_dropd = ['par_pri_emp', 'par_seg_emp', 'nov_emp_6m', 'def_emp_6m', 'qtd_sol_emp']
# sub3 = ['flag_telefone', 'flag_aadhar', 'flag_pan', 'flag_eleitor', 'flag_cmotorista', 'flag_passaporte']
# --------------------
# darei início à análise com as features: 'valor_emprestimo', 'custo_ativo', 'emprestimo_custo'
# suponho que estas possuam correlção entre si, sendo assim, cogito resumi-las em apenas uma variável,
# a presença de diversas features contendo informações sobre um mesmo aspecto acaba por atribuir um peso maior
# ao mesmo, além disso, não traz informações úteis e nos sujeitamos à maldição da dimensionalidade
# ao consultar a metadata:
# valor_emprestimo -> Valor total emprestado pela financeira
# custo_ativo -> Custo do ativo(veículo)
# emprestimo_custo -> Razão entre valor emprestado e valor do ativo(veículo)
# já 'por definição', conclui-se que emprestimo custo estará altamente correlacionada a uma combinação entre
# 'valor_emprestimo' e 'custo_ativo', que por sua vez, também possuem correlação entre si
# +
# ao observar o gráfico de dispersão entre as variáveis, percebemos a necessidade de eliminar outliers
# (ao menos para a visualização do gráfico de dispersão)
plt.scatter(df['valor_emprestimo'], df['custo_ativo'], alpha = 0.2)
plt.plot([0, 200000], [300000, 300000], color = 'grey', ls = ':')
plt.plot([200000, 200000], [0, 300000], color = 'grey', ls = ':')
plt.show()
aux = df.copy()
aux = aux.loc[(aux['valor_emprestimo'] <= 200000) & (aux['custo_ativo'] <= 300000)]
plt.scatter(aux['valor_emprestimo'], aux['custo_ativo'], alpha = 0.2)
plt.show()
# +
# precisa rodar duas vezes, r = 0.7387544522220792 (sem outliers), r = 0.756227463624271 (com outliers)
print('r:', pearsonr(df['valor_emprestimo'], df['custo_ativo']))
print('r:', pearsonr(aux['valor_emprestimo'], aux['custo_ativo']))
# eliminar outliers nos ajudou a visualizar melhor a correlação, no gráfico de dispersão, mas continuo
# o projeto sem eliminá-los
# +
# combinando 'valor_emprestimo' e 'custo_ativo'
# faz sentido fazer isso ? já que possuem correlação significativa, combino-as desta maneira
df['dif_val_custo'] = df['custo_ativo'] - df['valor_emprestimo']
# +
# agora nos restam as features 'dif_val_custo' e 'emprestimo_custo', que plotadas em um gráfico de dispersão,
plt.scatter(df['dif_val_custo'], df['emprestimo_custo'], alpha = 0.2)
plt.plot([175000, 175000], [10, 90], ls = ':', color = 'grey')
plt.show()
# novamente, outliers dificultam a visualização do plot
# estudo o cenário onde eliminamos os três pontos do lado direito da linha pontilhada cinza
aux = df.copy()
aux = aux.loc[aux['dif_val_custo'] <= 175000]
plt.scatter(aux['dif_val_custo'], aux['emprestimo_custo'], alpha = 0.2)
plt.show()
# +
# precisa rodar duas vezes r = -0.8429519408328675(com outliers), r = -0.8499209407779268 (sem outliers)
print('r:', pearsonr(df['dif_val_custo'], df['emprestimo_custo']))
print('r:', pearsonr(aux['dif_val_custo'], aux['emprestimo_custo']))
# eliminar outliers nos ajudou a visualizar melhor a correlação, no gráfico de dispersão, mas continuo
# o projeto sem eliminá-los
# não sei como cominar estas duas features, posto que uma consiste em uma proporção, e outra em um valor
# 'inteiro', então, avaliarei a importância de uma contra a outra, discrertizando-as com árvores de decisão,
# em seguida, plotando as taxas de 'default' para cada uma e julgando qual ordena o mesmo com maior eficácia
# +
# testo 16, 8 e 4 bins, mas o único número que traz resultados interessantes é 4 bins
# -
tree_box_plot(df, 'dif_val_custo', 'default', [4, 3, 2], 42)
# +
# conforme o eixo x cresce, maior a probabilidade de 'default', as ordenadas contém a variável quantitativa
# na próxima célula, realizo o mesmo processo em 'emprestimo_custo'
# -
tree_box_plot(df, 'emprestimo_custo', 'default', [4, 3, 2], 42)
# +
# no caso de 'emprestimo_custo', percebe-se que existe uma correlação significativa com a probabilidade de
# 'default'
# conclusão da análise de 'valor_emprestimo', 'custo_ativo', 'emprestimo_custo':
# talvez possamos usar a variável 'emprestimo_custo'
# +
# em seguida, faço a análise e transformação de 'nascimento'
# possíveis anos de nascimento:
# suponho que todos sejam '19--', com exceção de '1900', neste caso, considero que o ano seja '2000'
df['nascimento'].str[6:].unique()
# -
df['nascimento'] = df['nascimento'].str[6:].apply(lambda x: year_to_age(x))
df = df.rename(columns = {'nascimento': 'idade'})
tree_box_plot(df, 'idade', 'default', [4, 3, 2], 42)
# +
# conclusão da análise de idade:
# por mais que exista a possibilidade de gerar controvérsias, a variável idade ordena o 'default' de forma
# consistente
# +
# em seguida, faço a análise da variável emprego
print(df['emprego'].isnull().value_counts())
df['emprego'].unique()
# será necessário imputar alguns valores
# -
imp = SimpleImputer(strategy = 'constant', fill_value = 'Unknown')
df['emprego'] = imp.fit_transform(df[['emprego']])
df['emprego'] = df['emprego'].str.replace(' ', '_')
# +
# como se trata de uma variável qualitativa não ordinal, repetirei o processo das variáveis anteriores
# em vez disso, opto por realizar teste qui-quadrado, caso este acuse dependência, é realizado cramer V,
# mensurando a força da mesma
stat, crit, cramer = quali_var_indep_hyp_test(df, 'emprego', 'default', 0.01)
print(round(stat, 2))
print(round(crit, 2))
# -
# Com uma estatística qui-quadrada de 145.41, e um valor crítico de 9.21, falhamos em rejeitar a hipótese nula
# de independência, todavia, para quantificarmos a associação entre 'emprego' e 'default', consultamos
# Cramér's V, assim, sabemos se vale a pena manter esta variável no modelo
round(cramer, 3)
# +
# conclusão da análise de emprego:
# associação insignificante
# +
# em seguida, análise de 'score' e 'score_desc'
# suponho que seria seguro supor que estas variáveis trazem informações redundantes, entretanto, segue a
# análise destas, a fim de garantir que não perderemos informação, ao eliminar uma delas
# -
df['score_desc'].value_counts(1)
# +
# em um primeiro momento, de forma grosseira, considero a possibilidade de dividir os possíveis valores de
# 'score_desc' entre possuindo a palavra 'Risk' ou se iniciar com 'No'
# -
df.loc[df['score_desc'].str.contains('No'), 'score_desc'].unique().tolist()
df.loc[(df['score_desc'].str.contains('Risk')) | (df['score_desc'].str.contains('Risk')),
'score_desc'].unique().tolist()
# +
# à partir deste ponto, considero dois tipos de 'score_desc', os que contém 'Risk' e os que contém 'No'
# +
# como se comporta o 'score', em dados que possuem 'No' em 'score_desc' ?
# como é o score de dados nestes grupos ?
no_score = df.loc[df['score_desc'].str.contains('No'), 'score_desc'].unique().tolist()
no_score_score_series = df.loc[df['score_desc'].isin(no_score), 'score']
print('valor máximo:', no_score_score_series.max())
plt.hist(no_score_score_series, bins = 50)
plt.show()
# +
# dados que possuem 'No' em seus 'score_desc' consistem principalmente em zeros, chegando a, no máximo, 18,
# a princípio, não farei a discretização dos dados decidindo os limiares por conta própria, mas consigo
# imaginar que poderíamos alocar todos os dados contendo 'No' em 'score_desc' sob a mesma categoria
# +
# aprofundando análise dos valores de 'score_desc' que possuem 'Risk' ou 'risk'
print(df.loc[df['score_desc'].str.contains('Very Low'), 'score_desc'].unique())
print(df.loc[(df['score_desc'].str.contains('Low')) &
(df['score_desc'].str.contains('Very') == False), 'score_desc'].unique())
print(df.loc[df['score_desc'].str.contains('Medium'), 'score_desc'].unique())
print(df.loc[(df['score_desc'].str.contains('High')) &
(df['score_desc'].str.contains('Very') == False), 'score_desc'].unique())
print(df.loc[df['score_desc'].str.contains('Very High'), 'score_desc'].unique())
# +
# o que querem dizer as letras?
# r:são subgrupos de cada tipo de risco, o risco aumenta conforme se percorre o alfabeto
# Very Low Risk (A, B, C, D)
# Low Risk (E, F, G)
# Medium Risk (H, I)
# High Risk (J, K)
# Very High Risk (L, M)
# +
# como se comporta o 'score', para cada 'score_desc'
# dividir dados pela letra presente no 'score_desc'
df_risk_letter = df.copy()
df_risk_letter = df_risk_letter.loc[(df_risk_letter['score_desc'].str.contains('Risk')) |
(df_risk_letter['score_desc'].str.contains('risk'))]
df_risk_letter['risk_letter'] = df_risk_letter['score_desc'].str[0]
df_risk_letter[['risk_letter', 'score']].groupby('risk_letter').mean()
# +
# dividir dados pelo valor presente no 'score_desc', desconsiderando a letra
df_risk_word = df.copy()
df_risk_word = df_risk_word.loc[(df_risk_word['score_desc'].str.contains('Risk')) |
(df_risk_word['score_desc'].str.contains('risk'))]
df_risk_word['risk_word'] = df_risk_word['score_desc'].str[2:]
df_risk_word[['risk_word', 'score']].groupby('risk_word').mean().sort_values('score')
# +
# com estas informações, concluo que não há necessidade de manter ambas variáveis('score' e 'score_desc')
# opto por manter 'score'
# -
# prosseguindo com a análise de 'score'
tree_box_plot(df, 'score', 'default', [4, 3, 2], 42)
# resultado curioso, a expectativa era de que esta variável trouxesse muita informação sobre o alvo
# o que aconteceu em 3?
plt.hist(df.loc[df['score_proba'] == 3, 'score'])
plt.show()
# +
# o grande número de scores iguais a 0 tornam a variável score menos confiável,
# para contornar este problema, podemos alocar este grupo de dados sob a categoria 0, antes de passar os
# dados no modelo
# como ficariam os dados de 'score', sem o grande volume de zeros ?
aux = df.copy()
aux = aux.loc[df['score'] > 20]
tree_box_plot(aux, 'score', 'default', [4, 3, 2], 42)
# +
# notamos que com 8 ou mais bins, a variável continua inconsistente, mas com 4, ela passa a ser útil,
# após lidarmos com os zeros
# conclusão da análie de 'score' e 'score_desc' podemos utilizar 'score', alocando valores de 0-18 sob a
# categoria 0 e posteriormente, criando 4 bins para os valores restantes (seria melhor se todas as variáveis
# possuíssem o mesmo número de bins ?)
# +
# em seguida, análise de 'tem_med_emp' e 'tem_pri_emp'
# em que formato se encontram os dados ?
print(df['tem_med_emp'].unique()[0:5])
print(df['tem_pri_emp'].unique()[0:5])
# percebe-se que todos possuem inteiros, seguidos de 'yrs' e 'mon', (anos, meses)
# pretendo transformar os valores em meses apenas (1 ano = 12 meses)
df['tem_med_emp'] = df['tem_med_emp'].apply(lambda x: time_unit_unification(x))
df['tem_pri_emp'] = df['tem_pri_emp'].apply(lambda x: time_unit_unification(x))
# +
# tempo é uma grandeza contínua, sendo assim, lido com estas series da mesma forma que as variáveis contínuas
# já estudadas neste notebook
# proba_box(df, 'tem_med_emp', 'default', 3, 42)
# proba_box(df, 'tem_med_emp', 'default', 2, 42)
tree_box_plot(df, 'tem_med_emp', 'default', [4, 3, 2], 42)
# -
tree_box_plot(df, 'tem_pri_emp', 'default', [4, 3, 2], 42)
# +
# conclusão da análise de 'tem_med_emp' e 'tem_pri_emp':
# para considerar o uso de 'tem_med_emp' e 'tem_pri_emp', em ambos os casos, seria necessário trabalhar com
# 4 bins, se formos comparar os plots das variáveis, percebe-se uma tendência mais definida em 'tem_pri_emp'
# +
# em seguida, análise das variáveis referentes às contas primária e secundária: 'pri_qtd_tot_emp',
# 'pri_qtd_tot_emp_atv', 'pri_qtd_tot_def', 'pri_emp_abt', 'pri_emp_san', 'pri_emp_tom', 'sec_qtd_tot_emp',
# 'sec_qtd_tot_emp_atv', 'sec_qtd_tot_def', 'sec_emp_abt', 'sec_emp_san','sec_emp_tom'
# chegamos à conclusão de que podemos somar os dados das contas primária e secundária de cada dado
df['qtd_tot_emp'] = df['pri_qtd_tot_emp'] + df['sec_qtd_tot_emp']
df['qtd_tot_emp_atv'] = df['pri_qtd_tot_emp_atv'] + df['sec_qtd_tot_emp_atv']
df['qtd_tot_def'] = df['pri_qtd_tot_def'] + df['sec_qtd_tot_def']
df['emp_abt'] = df['pri_emp_abt'] + df['sec_emp_abt']
df['emp_san'] = df['pri_emp_san'] + df['sec_emp_san']
df['emp_tom'] = df['pri_emp_tom'] + df['sec_emp_tom']
# elas ordenam o 'default' ? são todas quantitativas
# -
acc_vars = ['qtd_tot_emp' ,'qtd_tot_emp_atv' ,'qtd_tot_def' ,'emp_abt' ,'emp_san' ,'emp_tom']
for var in acc_vars:
print(var)
proba_box(df, var, 'default', 3, 42)
proba_box(df, var, 'default', 2, 42)
# +
# depois desta análise, acho que talvez tenha feito algo errado ou que esteja interpretando mal os dados
# o fato de os ranges que compõe cada categoria criada na discretização serem maiores ou menores quer dizer que
# é mais ou menos interessante usar a variável ? ou todas valem o mesmo, desde que exista ordem ?
# conclusão da análise das variáveis referentes às contas:
# todos os resultados são decepcionantes
# +
# neste ponto do notebook, todas as features que nos propusemos a analisar foram analisadas
# -
# as features que se mostraram mais interessantes foram 'emprestimo_custo_proba' , 'idade_proba',
# 'score_proba' ,'tem_pri_emp_proba', em 4 bins (score precisa ser tratada à parte)
df_x = df.copy()
df_x = df_x[['emprestimo_custo_proba', 'idade_proba', 'tem_pri_emp_proba', 'score', 'default']]
df_x.loc[df_x.index.isin(df_x.loc[df_x['score'] < 20].index), 'score'] = 0
x = df_x.loc[df_x['score'] != 0][['score']]
y = df_x.loc[df_x['score'] != 0][['default']]
mdl = DecisionTreeClassifier(random_state = 42, max_depth = 2)
mdl.fit(x, y)
fig = plt.figure(figsize = (12, 5))
_ = tree.plot_tree(mdl)
plt.show()
# +
def score_new_disc(x):
if x <= 438.5:
return 1
elif x > 438.5 and x <= 615.5:
return 2
elif x > 615.5 and x <= 738.5:
return 3
elif x > 738.5:
return 4
z_series = df_x.loc[df_x['score'] == 0]['score']
nonz_series = df_x.loc[df_x['score'] != 0]['score'].apply(lambda x: score_new_disc(x))
disctzd_score = pd.concat([z_series, nonz_series])
disctzd_score = disctzd_score.rename('a')
df_x = df_x.join(disctzd_score).drop(columns = ['score'])
# -
target_prop_bin(df_x, 'a', 'default')
target_prop_bin(df_x, 'idade_proba', 'default')
target_prop_bin(df_x, 'emprestimo_custo_proba', 'default')
target_prop_bin(df_x, 'tem_pri_emp_proba', 'default')
# +
# x = df_x[['emprestimo_custo_proba', 'idade_proba', 'tem_pri_emp_proba', 'a']]
# y = df_x['default']
# lr = LogisticRegression()
# lr.fit(x, y)
# lista = [i[1] for i in lr.predict_proba(x)]
# roc_auc_score(y, lr.decision_function(x))
# roc_auc_score(y, lista)
# (2 * roc_auc_score(y, lista)) - 1
| 19,778 |
/wavenet_store_info_featureEng .ipynb | fab82e58deb387bc604188cc8395e4a7bbfbab76 | [] | no_license | GatoY/g-forecast | https://github.com/GatoY/g-forecast | 2 | 0 | null | null | null | null | Jupyter Notebook | false | false | .py | 62,551 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# + deletable=false editable=false run_control={"frozen": true}
# # Lecture 2 - Python Input, Printing, Scripts, and Functions
# ---
# -
# Replace `Your name here` in the following cell with your first and last name then execute the cell
name = "Your name here"
print("Name:", name.upper())
# + deletable=false editable=false run_control={"frozen": true}
# Execute the time stamp cell below
# + deletable=false editable=false
from datetime import datetime
from pytz import timezone
print(datetime.now(timezone('US/Eastern')))
# -
# ## Purpose
#
# - Use the `.format()` string method to generate specifically formatted output
#
# - Use the `input()` function to generate interactive input from the user in scripts and user-defined functions
#
# - Create, edit, and execute simple scripts using *Python*
#
# - Assign values to variable names within scripts
#
# - Request user input to assign values to variables in scripts using the `input()` function
#
# - Create and execute user-defined functions that do and do not accept arguments
#
# - Create and execute void and fruitful functions
#
# - Use `print()` to display output from scripts and user-defined functions
#
# ## Instructions
#
# 1. Replace `Your name here` in the cell below the assignment title with your first and last name and then execute the cell using "Shift-Enter"
# 2. Execute the time stamp cell using "Shift-Enter"
# 3. Follow along with the instructor in class as we use *Python* to generate formatted output, request interactive user input, and create scripts and functions
# 4. Execute the date stamp cell at the end of the document and save the file
# + deletable=false editable=false run_control={"frozen": true}
# ## Some Creative Commons Reference Sources for This Material
#
# - *Think Python 2nd Edition*, Allen Downey, chapters 3 and 6
# - *The Coder's Apprentice*, Pieter Spronck, chapters 5 and 8
# - *A Practical Introduction to Python Programming*, Brian Heinold, chapters 1, 10, 13, and 23
# - *Algorithmic Problem Solving with Python*, John Schneider, Shira Broschat, and Jess Dahmen, chapters 3 and 4
# + deletable=false editable=false run_control={"frozen": true}
# ## Reviewing the `print()` Function
#
# Recall that *Python's* `print()` function can be used to display numeric values and text strings. Multiple items can be printed using the same `print()` function by separating items with commas. *Python* adds a space between items that are separated by a comma. You can force a line return in any string by adding the newline escape sequence `\n`. There are other escape sequences as well. A good one to remember is `\t` for adding a tab. You will notice that multiplying a string by an integer will print the string that number of times.
#
# + deletable=false editable=false run_control={"frozen": true}
# ___
# **Practice it**
#
# Type and execute the following `print()` expressions in the following code cell.
#
# ```python
# print("Python is awesome")
# print('Lumberjacks', "Parrots", 42)
# print(4*5)
# print(2*'Hello')
# print('Hello,\nWorld!')
# ```
# -
# + deletable=false editable=false run_control={"frozen": true}
# ## Functions Versus Methods
#
# *Python* uses both functions and methods to work with/on objects. Methods are a lot like functions in that they both accept arguments. Their syntax is different though, as is shown in the provided images. Functions usually work with arguments to return a value or do something (like the `abs()` or `print()` functions). Methods usually work on an object using arguments to either return a value or change the object. We will look at the string `.format()` method today.
#
# 
#
# 
# + deletable=false editable=false run_control={"frozen": true}
# ## Formatting Printed Output
#
# ### The `.format()` String Method
#
# If we include the `.format()` string method within a `print()` function, we can control exactly how numeric (and non-numeric) values are displayed. The general layout of the `.format()` method for a string is as follows:
#
# `'The sum of {} and {} is {}'.format(item_1, item_2, item_3)`
#
# The expression starts with a string that has curly braces `{}` as placeholders for numeric or string objects. The `.format()` method directly follows the closing quote for the string. The arguments located between the method's parentheses are the values (in order) that match the placeholders in the string. These may be values and/or expressions. The placeholders themselves can include formatting descriptors to generate very specific formatting, especially for numeric values. Formatting descriptors within braces must be preceded by a colon, i.e. `{:.2f}`.
#
# The website https://pyformat.info does a good job of explaining a number of varied examples of the `.format()` method.
#
# + deletable=false editable=false run_control={"frozen": true}
# ___
# **Practice it**
#
# Display the result of `22/7` (an estimate for $\pi$ that has historically been used by many tool makers and machinists) in a variety of ways using `print()` and the `.format()` string method. Modify the formatting descriptors within the curly braces in each of the following code cells then execute them.
#
# - Using `print()` with two arguments and a comma but no formatting
# + deletable=false editable=false run_control={"frozen": true}
# # Standard print() without .format()
# print('pi is close to', 22/7)
# + deletable=false editable=false run_control={"frozen": true}
# - `{}` No specific formatting assigned
# + slideshow={"slide_type": "fragment"}
print('pi is close to {}'.format(22/7))
# + deletable=false editable=false run_control={"frozen": true} slideshow={"slide_type": "fragment"}
# - `{:f}` Standard floating point notation with the default number of decimal places
# + slideshow={"slide_type": "fragment"}
print('pi is close to {}'.format(22/7))
# + deletable=false editable=false run_control={"frozen": true}
# ### Formatted String Literals (Python 3.6+)
#
# New to *Python* starting with version 3.6 are *formatted string literals* or "*f-strings*". These work like the `.format()` method but in a more direct way. For example, instead of using `print('pi is close to {}'.format(22/7))` you can use `print(f'pi is close to {22/7}')`. Formatted string literals allow expressions or variables to be placed directly within the curly braces. If special formatting is desired for a value, a colon is added after the expression of variable with the formatting after the colon, i.e. `print(f'pi is close to {22/7:.8f}')`.
#
# ___
# **Practice it**
#
# Modify the formatting descriptors within the curly braces in each of the following code cells then execute them.
#
# - `{:f}` Standard floating point notation with the default number of decimal places
#
# -
print(f'pi is close to {22/7}')
# + deletable=false editable=false run_control={"frozen": true}
# - `{:.16f}` Standard floating point notation with 16-decimal places
# -
print(f'pi is close to {22/7}')
# + deletable=false editable=false run_control={"frozen": true} slideshow={"slide_type": "fragment"}
# - `{:e}` Exponential notation (use `E` for uppercase)
# -
print(f'pi is close to {22/7}')
# + deletable=false editable=false run_control={"frozen": true} slideshow={"slide_type": "fragment"}
# - `{:.12E}` Exponential notation with 12-decimal places
# -
print(f'pi is close to {22/7}')
# + deletable=false editable=false run_control={"frozen": true} slideshow={"slide_type": "fragment"}
# - `{:g}` Standard or exponential notation, whichever is more efficient (use `G` for uppercase)
# -
print(f'pi is close to {22/7}')
# + deletable=false editable=false run_control={"frozen": true} slideshow={"slide_type": "fragment"}
# - `{:.12g}` 12-decimal places and automatically use standard or exponential notation, whichever is shorter
# -
print(f'pi is close to {22/7}')
# + deletable=false editable=false run_control={"frozen": true} slideshow={"slide_type": "fragment"}
# - `{:8.3f}` Total width to 8 characters with 3 to the right of the decimal
# -
print(f'pi is close to {22/7}')
# + deletable=false editable=false run_control={"frozen": true}
# *What does this formatting descriptor mean?* The `8` means that there are 8 total characters set aside for displaying the number, the `.3` says there are to be 3 digits to the right of the decimal point, and `f` means the value will be formatted as a floating point value.
#
# ```
# | | | |3|.|1|4|3| <= formatted value
# | | | | | | | | |
# |8|7|6|5|4|3|2|1| <= characters set aside for the value
# ```
# Notice that the decimal point counts as a character. There are 8 total characters set aside to display the value with 3 of them to the right of the decimal point. If all 8 characters are not needed for the value, then *Python* will add spaces to the left in order to use all 8 characters.
# + deletable=false editable=false run_control={"frozen": true}
# - `{:+08.3f}` Same as the previous but with leading zeros and the +/- sign
# -
print(f'pi is close to {22/7}')
# + deletable=false editable=false run_control={"frozen": true}
# - `{:.0f}` Force a floating point to display no values right of the decimal
# -
print(f'pi is close to {22/7}')
# + deletable=false editable=false run_control={"frozen": true}
# - You can use varables withing f-strings as well as values and calculations. Try it by executing the following cell after setting the formatting to 8 decimal places.
# -
almost_pi = 22/7
print(f'pi is close to {almost_pi}')
# + deletable=false editable=false run_control={"frozen": true}
# ___
# **Practice it some more**
#
# Here are few more common and useful formatting descriptors. Modify and execute each of them to see the output they generate.
#
# - `{:d}` Standard integer (object must be of `int` type)
# -
print(f'Integer value: {42}')
# + deletable=false editable=false run_control={"frozen": true}
# - `{:4d}` Integer with 4 total characters
# -
print(f'Integer value: {42}')
# + deletable=false editable=false run_control={"frozen": true}
# - `{:04d}` Integer with 4 total characters and leading zeros
# -
print(f'Integer value: {42}')
# + deletable=false editable=false run_control={"frozen": true}
# - `{:+04d}` Integer with 4 total characters, leading zeros, and the +/- sign
# -
print(f'Integer value: {42}')
# + deletable=false editable=false run_control={"frozen": true}
# - `{:,d}` Integer with comma separators
# -
print(f'Integer value: {987654321}')
# + deletable=false editable=false run_control={"frozen": true}
# - `{:s}` String (although setting the formatting is not really necessary in this case)
# -
first_name = "Slim"
last_name = "Shady"
print(f'I am the real {first_name} {last_name}')
# + deletable=false editable=false run_control={"frozen": true}
# ## The `input()` Function
#
# The `input()` function is used to request information from a user at a command line so it can be used in a script. It accepts one optional argument; a statement or question to the user so they know what to enter. The argument must be a string. This function always returns whatever the user has typed as a string. You need to specifically convert the returned value to an integer or float if that is what you actually want. The following examples illustrate typical usages.
#
# ```python
# user_name = input('What is your name? ')
# city = input("Enter your city of residence: ")
# applied_load = float(input('Enter the applied load (lbf): '))
# age = int(input('Enter your current age > '))
# ```
#
# Notice that the last two examples have the `input()` function inside of `float()` and `int()` type conversion functions. Since strings are always returned from `input()` functions, you need to convert the result to the type of value desired before performing any calculations. This is an easy way to do it.
#
# It is considered good practice to end `input()` function prompt strings with a delimiter of some sort and a space. These examples use common delimiters; question mark (`?`), colon (`:`), and a right arrow (`>`). The delimiter and space helps separate the prompt from the response and makes it easier to read. This is something you should do when writing your own prompts.
# + deletable=false editable=false run_control={"frozen": true}
# ___
# **Practice it**
#
# Write and execute three `input()` functions with prompts and variable assignments.
#
# 1. Request text-based information
# 2. Ask for an integer and convert the response
# 3. Ask for a decimal value and convert the response appropriately as well
#
# Make sure you end each prompt string with a delimiter and space.
# -
# + deletable=false editable=false run_control={"frozen": true}
# ## Scripting and Functions Background
#
# Anything that can be done with *Python* from the REPL or in a *Jupyter* notebook can also be done via a script or user-defined function (more efficiently most of the time). Scripts, sometimes called programs, in their simplest form are essentially lists of commands that are executed sequentially (one after another) from top to bottom. The variables used in scripts can be assigned values a number of ways, although only the first two will be explored at this time:
#
# 1. Assign within the script (hard-coded)
# 2. Ask the user to input a value at a prompt
# 3. Pass when the script is executed
# 4. Load values from a file
# + deletable=false editable=false run_control={"frozen": true}
# User-defined functions are similar to scripts except they generally receive input by passing arguments instead of using `input()`. For example, when we previously used the `abs()` function, we would pass a value to it for which we wanted to get the absolute value. The expression `abs(-100)` passes `-100` as an argument to the absolute value function. Functions can also be included inside a script or as part of a module. If a function is defined within a script, it must be done before the function is called (used) in the script. Good programming practice says to place all `import` statements at the beginning of scripts and all user-defined functions immediately thereafter.
#
# Once a variable name is assigned in a *Python* script it is available for use in commands further down in the script. On the other hand, variables passed into and used in a function are only available to use inside the function. The results generated from performing calculations in scripts can be displayed/output either using the `print()` function or by writing output values to a text file, although only the first will be used at this time. User-defined functions utilize these techniques as well, but more often they just return results back to the calling location. For example, `abs(-100)` returns the value `100`. Not all user-defined functions return values. Those that do not are sometimes called **void functions** and those that do are sometimes called **fruitful functions**.
#
# Unlike when strictly working at the command line or from a *Jupyter* notebook interface, you cannot correct for typos, re-execute an expression, and keep going when running a script. You, the programmer, will have to make sure that all of the commands in the script are error free and the script provides the necessary and correct output in an understandable manner. Script files and user-defined functions should include a liberal dose of comments so the programmer and others can understand why things are being done a particular way in the event that changes need to be made in the future.
# + deletable=false editable=false run_control={"frozen": true}
# ## Creating, Editing, and Executing a Script
#
# Scripts are written as plain text files using text editors. The *Jupyter* environment includes a simple text editor that has syntax highlighting to color commands, functions, values, strings, and other objects. *Python* script files end with a `.py` extension. Executing *Python* scripts can done from a standard command line prompt by typing `python` or `python3` (depending upon the version being used and/or how it is installed) followed by a space and the script name including the extension. For example, executing a script named `my_script.py` with *Python 3.x* is accomplished by typing `python3 script_name.py` and pressing the `[enter]` key.
#
# If the script file is located in the same folder/directory as a *Jupyter* notebook, you can execute the script in the notebook environment. To do so for the script named `my_script.py`, execute the expression `run my_script.py` from a code cell. The same technique can be used from within the *iPython* command line environment.
#
# Many scripts (probably all of them created in this class) can be executed at a *Python* prompt by importing it. For example, `import my_script` will execute the script named `my_script.py`.
#
# It is good practice to include comments at the top of a script file that describes what the script does, units that are used, special conditions, etc. Many people also like to include a comment that includes their name, website, licensing, etc.
# + deletable=false editable=false run_control={"frozen": true}
# ___
# **Practice it: Creating Your First Script File**
#
# Create a **New** text file named `mph2kph.py`. It is important that you don't have any spaces before or in the script name and that you include the extension. Use the editor to create a short script as outlined below that converts from mph to km/h. Start by copying the provided outline to the new script. "Hard code" the value of `mph` instead of using an `input()` for this first script.
#
# Execute your script from this notebook when done by typing `run mph2kph.py` in the provided code cell.
# + active=""
# #==========================================================
# # this file converts mph to km/h
# # the conversion factor is 1 mph = 1.609 km/h
# #
# # author: brian brady (use your name)
# # class: MECH 322 (8am, 10am, 3pm)
# #==========================================================
#
# # assign a value to a variable named 'mph'
#
# # assign the value 1.609 to a variable named 'conversion'
#
# # calculate and assign the speed in km/h to the variable 'kph'
#
# # use a print() function to create output similar the following example:
# # 60 mph is 96.53999999 km/h
#
#
# # end of script
# -
# Run you script here (replace this entire line of text with the command to run the script)
# + deletable=false editable=false run_control={"frozen": true}
# ___
# **Practice it**
#
# Make a duplicate the previous script and name it `mph2kph_input.py`. Instead of assigning a fixed value to `mph`, use an `input()` function that asks the user to enter a speed in mph. You will need to place the `input()` function inside the parentheses of a `float()` function to convert the input value so you can perform calculations with the value. Change the `print()` expression to use formatted output and displays the input speed with no special formatting and the calculated speed with one decimal place. Run the script in the three empty code cells below using three different speeds of your choosing.
# -
# + deletable=false editable=false run_control={"frozen": true}
# ___
# **Practice it**
#
# Create a script called `windchill.py` that uses `input()` functions to ask the user to enter the air temperature in degrees F and the wind speed in mph then calculates the wind chill temperature $T_{wc}$ in degrees F.
#
# $\qquad\displaystyle T_{wc}=35.74 + 0.6215 \,T - 35.75 \,v^{0.16} + 0.4275\,T \,v^{0.16} $
#
# Print the statement **"xxx degrees F with a wind speed of yyy mph equals a wind chill of zzz degrees F"** using formatted printing such that the input values have no special formatting and the calculated value is a float with zero decimal places. Include introductory comments with this script that describes what it does, the units being used, and your name (similar to the previous scripts). Test the script three times with the following values:
#
# - $30^{\circ}\text{F}$ with $10 \text{ mph}$
# - $10^{\circ}\text{F}$ with $30 \text{ mph}$
# - $-10^{\circ}\text{F}$ with $20 \text{ mph}$
#
# You should get results of $21^{\circ}\text{ F}$, $-12^{\circ}\text{ F}$, and $-35^{\circ}\text{ F}$.
#
# -
# + deletable=false editable=false run_control={"frozen": true}
# ## Creating User-Defined Functions
#
# Notice that in the above script example all of the lines were aligned on the left edge. This is very important when writing scripts in *Python* as indentation is used to group commands for specific purposes. Therefore, the default indentation is no indentation at all. Even a single space at the beginning of a line will generate an error; test it by executing the following cell.
# -
# The following line has a space at the beginning
print("Spaces are important")
# + deletable=false editable=false run_control={"frozen": true}
# When creating a user-defined function, we will need to indent all commands that belong to the function except the first line in order to signal to *Python* that they belong together. The following image illustrates the basic structure of user-defined functions:
#
# 
# + deletable=false editable=false run_control={"frozen": true}
# The first line of the above function definition is called the **function header** and includes a function name followed by names of any arguments (if there are any) that need to be passed into the function. The end of the function header line *must* end with a colon. All other lines must be indented by 4 spaces and are called the **function body**. The body must consist of at least one command. If nothing at all is being done by the function, then the `pass` command must be used. When *Python* reads the `pass` command, it immediately exits the function and returns to where it was. Function definitions may optionally include a **docstring** that is used to display information about the function when `help()` is called with the function name as an argument. The docstring must be enclosed by three double quotes and can span multiple lines.
# + deletable=false editable=false run_control={"frozen": true}
# ### Void Functions
#
# Functions that do not return any values can be considered to be **void**. They may perform a calculation and print the results, but they don't pass anything back to where they were called. The above function definition is for a void function. In fact, it is a useless void function since it only contains a `pass` command.
#
# + deletable=false editable=false run_control={"frozen": true}
# ___
# **Practice it**
#
# Below is a function definition for a void function that simply prints `Hello, World!`. Notice that it does not accept any arguments (the parentheses in the header are empty). Execute the code cell to place the function in memory. Then call the function in the next code cell using `hello()` (make sure that you include the parentheses). Ask for help on the function in the second code cell.
# -
def hello():
"""This function just prints the phrase 'Hello, World!'
It does not return any results and it does not accept any arguments
"""
print('Hello, World!')
# + deletable=false editable=false run_control={"frozen": true}
# ___
# **Practice it**
#
# The following code block contains another void function that accepts a single argument and prints the result of the argument value multiplied by $2$. Execute the code cell with the function definition and then execute the function with any numeric argument of your choosing in the next code cell.
# -
def double_me(value):
"""This function multiplies the argument named value by 2
and prints the result
"""
doubled_value = value * 2
print(doubled_value)
# + deletable=false editable=false run_control={"frozen": true}
# ___
# **Practice it**
#
# This next example is also a void function, but it accepts two arguments. Add an expression in the `print()` function parentheses to multiply the first argument by the second and then execute the code cell to place it in memory. Execute the function in the blank code cell with any pair of numeric values. Notice that the argument names used in this and the previous example are not the same. You can use any valid variable name as a function argument name.
# -
def multiply2(arg1, arg2):
print()
# + deletable=false editable=false run_control={"frozen": true}
# ___
# **Practice it**
#
# In the following code cell define a function called `print_mph2kph` that takes one argument named `mph`. The function should convert `mph` to km/h and assign the result to the name `kph`. On the last line of the function print the result so that it reads **"xxx mph equals yyy km/h"**, where **"xxx"** and **"yyy"** are replaced by values of `mph` and `kph`. In the next two empty code cells test the function with two different speeds.
# -
# + deletable=false editable=false run_control={"frozen": true}
# ### Fruitful Functions
#
# Fruitful functions return a value back to the caller and usually do not create any printed output. In order to return a value (or values) fruitful functions need to have a `return` statement. The `return` is typically located on the last line of a function since *Python* exits the function as soon as a value is returned. More than one value can be returned from a function by including multiple values separated by commas after the `return` statement. Keep in mind that unlike the `print()` function, `return` is a statement and does not use parentheses. The example below illustrates the general structure of a fruitful function.
#
# ``` python
# def my_function(arg1,arg2):
# """docstring"""
# body line 1
# body line 2
# return value1, value2
# ```
# + deletable=false editable=false run_control={"frozen": true}
# ___
# **Practice it**
#
# The following incomplete function definition is a modification of the `double_me` function called `double_me2`. This time, instead of printing the result of the calculation, the function should `return` the result. Modify the function definition so that it returns `doubled_value`. Execute the code cell to place the function into memory and then test the function in the next two blank code cells with values of your choosing. Assign the result of the function call to the variable name `two_times` in the second test cell and then print `two_times`. Do you remember how to assign a variable name to a calculation?
# -
def double_me2(value):
"""This function multiplies the argument named value by 2
and returns the result
"""
doubled_value = value * 2
# + deletable=false editable=false run_control={"frozen": true}
# A more **Pythonic** way to define the above function would be to move the calculation to the `return` line. Unless the intermediate variable is needed for another calculation in the function, it is more efficient to just return the calculation directly.
#
# ___
# **Practice it**
#
# Edit the following function definition to perform the calculation on the `return` line and execute the function definition to place it into memory. Then call this new function with a value of your choosing.
# -
def double_me3(value):
"""This function multiplies the argument named value by 2
and returns the result
"""
# + deletable=false editable=false run_control={"frozen": true}
# ___
# **Practice it**
#
# The following (incomplete) function named `rectangles` should return the area and perimeter of a rectangle with sides of `width` and `height`. Edit the function definition such that the `return` line includes the calculations for area and perimeter. Execute the function definition then test the function with two different sets of arguments of your choosing.
#
# For the second test, assign the function call to a set of varibles like so, `(a, p) = rectangles(width, height)` except with your numeric width and height values. The grouping `(a, p)` is referred to as a **tuple** by *Python* and is the *Pythonic* way of assigning multiple values to varaible names at the same time. In the final blank code cell print `a` and `p`.
# -
def rectangles(width, height):
"""Returns the area and perimeter (in that order) for a rectangle of width and height"""
return
# + deletable=false editable=false run_control={"frozen": true}
# ___
# **Practice it**
#
# Copy the previously created `print_mph2kph` function into the following code cell. Rename this version of the function to `return_mph2kph` and modify the function body such that it returns the speed in km/h but does not print anything. Test this version with the same speeds as above in the provided code cells. In the second code cell, assign the result of the function call to the name `speed` and then `print(speed)`.
# -
# + deletable=false editable=false run_control={"frozen": true}
# ___
# **Practice it**
#
# Define a function called `windchill` that does the same thing as the script from earlier called `windchill.py` except instead of using `input()` functions to get the air temperature and wind speed it uses two arguments called `tempF` and `vel_mph`. Round the resulting wind chill temperature to zero decimal places. Have the function both print the statement **"xxx degrees F with a wind speed of yyy mph equals a wind chill of zzz degrees F"** and return the wind chill temperature. Include a docstring with this function.
#
# $\qquad\displaystyle T_{wc}=35.74 + 0.6215 \,T - 35.75 \,v^{0.16} + 0.4275\,T \,v^{0.16} $
#
# Test the function three times with the following values:
# - $30^{\circ}\text{F}$ with $30 \text{ mph}$
# - $20^{\circ}\text{F}$ with $20 \text{ mph}$
# - $10^{\circ}\text{F}$ with $10 \text{ mph}$
#
# You should get results of $15^{\circ}\text{F}$, $4^{\circ}\text{F}$, and $-4^{\circ}\text{F}$.
# -
# + deletable=false editable=false run_control={"frozen": true}
# ___
# **Wrap it up**
#
# Execute the code cell below to create a time and date stamp.
#
# Click on the **Save** button and then **Close and halt** from the **File** menu when you are done. **This is an instructor-led assignment that must be completed before the end of the lab session in order to receive credit.**
# + deletable=false editable=false
from datetime import datetime
from pytz import timezone
print(datetime.now(timezone('US/Eastern')))
| 30,597 |
/Week 3 - Quantum Gates/exercises/w3_01_s.ipynb | 789947c4670bd1c1b218a34295bce3cfe436701a | [] | no_license | osbaldoisaias/Teach-Me-Quantum | https://github.com/osbaldoisaias/Teach-Me-Quantum | 0 | 0 | null | null | null | null | Jupyter Notebook | false | false | .py | 58,896 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Quantum Gates in Qiskit
# Start by some typical setup and definition of useful functions, which you are encouraged to look at.
#
# Then, head to the [exercises start](#Exercises-Start-Here) to start coding!
from qiskit import QuantumCircuit, ClassicalRegister, QuantumRegister
from qiskit import execute
# Choose the drawer you like best:
from qiskit.tools.visualization import matplotlib_circuit_drawer as draw
#from qiskit.tools.visualization import circuit_drawer as draw
from qiskit import IBMQ
IBMQ.load_accounts() # make sure you have setup your token locally to use this
# %matplotlib inline
# ## Utils for visualizing experimental results
# +
import matplotlib.pyplot as plt
def show_results(D):
# D is a dictionary with classical bits as keys and count as value
# example: D = {'000': 497, '001': 527}
plt.bar(range(len(D)), list(D.values()), align='center')
plt.xticks(range(len(D)), list(D.keys()))
plt.show()
# -
# ## Utils for executing circuits
from qiskit import Aer
# See a list of available local simulators
print("Aer backends: ", Aer.backends())
# see a list of available remote backends (these are freely given by IBM)
print("IBMQ Backends: ", IBMQ.backends())
# ### Execute locally
# execute circuit and either display a histogram of the results
def execute_locally(qc, draw_circuit=False):
# Compile and run the Quantum circuit on a simulator backend
backend_sim = Aer.get_backend('qasm_simulator')
job_sim = execute(qc, backend_sim)
result_sim = job_sim.result()
result_counts = result_sim.get_counts(qc)
# Print the results
print("simulation: ", result_sim, result_counts)
if draw_circuit: # draw the circuit
draw(qc)
else: # or show the results
show_results(result_counts)
# ### Execute remotely
from qiskit.providers.ibmq import least_busy
import time
# Compile and run on a real device backend
def execute_remotely(qc, draw_circuit=False):
if draw_circuit: # draw the circuit
draw(qc)
try:
# select least busy available device and execute.
least_busy_device = least_busy(IBMQ.backends(simulator=False))
print("Running on current least busy device: ", least_busy_device)
# running the job
job_exp = execute(qc, backend=least_busy_device, shots=1024, max_credits=10)
lapse, interval = 0, 10
while job_exp.status().name != 'DONE':
print('Status @ {} seconds'.format(interval * lapse))
print(job_exp.status())
time.sleep(interval)
lapse += 1
print(job_exp.status())
exp_result = job_exp.result()
result_counts = exp_result.get_counts(qc)
# Show the results
print("experiment: ", exp_result, result_counts)
if not draw_circuit: # show the results
show_results(result_counts)
except:
print("All devices are currently unavailable.")
# ## Building the circuit
def new_circuit(size):
# Create a Quantum Register with size qubits
qr = QuantumRegister(size)
# Create a Classical Register with size bits
cr = ClassicalRegister(size)
# Create a Quantum Circuit acting on the qr and cr register
return qr, cr, QuantumCircuit(qr, cr)
# ---
# <h1 align="center">Exercises Start Here</h1>
#
# Make sure you ran all the above cells in order, as the following exercises use functions defined and imported above.
# ## Adding Gates
# ### Hadamard
# This gate is required to make superpositions.
#
# **TASK:** Create a new circuit with 2 qubits using `new_circuit` (very useful to reconstruct your circuit in Jupyter)
qr, cr, circuit = new_circuit(2)
# **TASK:** Add a Hadamard on the _least important_ qubit
# H gate on qubit 0
circuit.h(qr[0]);
# **TASK:** Perform a measurement on that qubit to the first bit in the register
# measure the specific qubit
circuit.measure(qr[0], cr[0]); # ; hides the output
# **TASK:** check the result using `execute_locally` test both `True` and `False` for the `draw_circuit` option
# Try both commands:
execute_locally(circuit,draw_circuit=True)
# execute_locally(circuit,draw_circuit=False)
# The result should be something like `COMPLETED {'00': 516, '01': 508}`.
#
# **TASK:** What does this mean?
# > That we got our superposition as expected, approximately 50% of the experiments yielded 0 and the other 50% yielded 1.
# ---
# ### X Gate (Pauli-X)
# This gate is also referred to as a bit-flip.
#
#
# **TASK:** Create a new circuit with 2 qubits using `new_circuit` (very useful to reconstruct your circuit in Jupyter)
qr, cr, circuit = new_circuit(2)
# **TASK:** Add an X gate on the _most important_ qubit
# H gate on qubit 1
circuit.x(qr[1]);
# **TASK:** Perform a measurement on that qubit to the first bit in the register
# measure the specific qubit
circuit.measure(qr[1], cr[0]); # ; hides the output
# **TASK:** check the result using `execute_locally` test both `True` and `False` for the `draw_circuit` option
# Try both commands:
execute_locally(circuit,draw_circuit=True)
# execute_circuit(circuit,draw_circuit=False)
# ## Free flow
# At this stage you are encouraged to repeat (and tweek as you wish) the above tasks for the Hadamard and X gates, especially on single qubit gates.
# ---
# ### CNOT (Controlled NOT, Controlled X gate)
# This gate uses a control qubit and a target qubit to
#
#
# **TASK:** Create a new circuit with 2 qubits using `new_circuit` (very useful to reconstruct your circuit in Jupyter)
qr, cr, circuit = new_circuit(2)
# **TASK:** Add a CNOT gate with the _least important_ qubit as the control and the other as the target
# CNOT gate
circuit.cx(qr[0], qr[1]);
# **TASK:** Perform a measurement on the qubits
# measure the specific qubit
circuit.measure(qr, cr); # ; hides the output
# **TASK:** check the result using `execute_locally` test both `True` and `False` for the `draw_circuit` option
# Try both commands:
execute_locally(circuit,draw_circuit=True)
# execute_circuit(circuit,draw_circuit=False)
# **TASK:** Since a single CNOT does not seem very powerful, go ahead and add a hadamard gate to the two qubits (before the CNOT gate) and redo the experiment (you can try this by using a single Hadamard on each qubit as well).
qr, cr, circuit = new_circuit(2)
# H gate on 2 qubits
circuit.h(qr);
# CNOT gate
circuit.cx(qr[0], qr[1]);
# measure
circuit.measure(qr, cr); # ; hides the output
# Try both commands:
execute_locally(circuit,draw_circuit=True)
# execute_circuit(circuit,draw_circuit=False)
# ## Free flow: Changing the direction of a CNOT gate
# Check this [application of the CNOT](https://github.com/Qiskit/ibmqx-user-guides/blob/master/rst/full-user-guide/004-Quantum_Algorithms/061-Basic_Circuit_Identities_and_Larger_Circuits.rst#changing-the-direction-of-a-cnot-gate) and try to replicate it using Qiskit!
#
# Try to replicate it using the unitary transformations as well, pen and paper is better suited for this.
#
# 
#
# A CNOT equals Hadamards on both qubits an oposite CNOT and two new Hadamards!
# ## Free flow: Swapping the states of qubits with a CNOT gate
# Check this [application of the CNOT](https://github.com/Qiskit/ibmqx-user-guides/blob/master/rst/full-user-guide/004-Quantum_Algorithms/061-Basic_Circuit_Identities_and_Larger_Circuits.rst#swapping-the-states-of-qubits) and try to replicate it using Qiskit!
#
# Try to replicate it using the unitary transformations as well, pen and paper is better suited for this.
#
# 
#
# Three CNOT gates allow 2 qubits to swap their original values, can you do this with 2 classical bits??
# ## Executing on a remote device
# If you do this, you may have to wait for some time (usually a few minutes), depending on the current demand of the devices
#
# **TASK:** Create a circuit that simply measures 5 qubits and run it on a remote device using `execute_remotely`!
qr, cr, circuit = new_circuit(5)
circuit.measure(qr, cr);
execute_remotely(circuit)
# **TASK:** Comment on the results
# >
# **Important:** Once you get the results, you may see that, in fact, most of the iterations resulted in `00000`, but you will also see that there will be a few hits on other bit configurations (typically mostly composed of `0`s, like `00001` or `00010`) this is due to **experimental error** on the quantum device and is a concern to take into account when deploying into real devices!!
| 8,778 |
/ASR/2_command_recognition/01-speech-commands-mfcc-extraction.ipynb | 551ee7d7a0f8452c287f1822a5dfeb03604d59b0 | [] | no_license | SusmoyBarman1/Jarvis | https://github.com/SusmoyBarman1/Jarvis | 0 | 0 | null | null | null | null | Jupyter Notebook | false | false | .py | 45,946 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
from os import listdir
from os.path import isdir, join
import librosa
import random
import numpy as np
import matplotlib.pyplot as plt
import python_speech_features
# Dataset path and view possible targets
dataset_path = 'C:\\Users\\SuSu\\Desktop\\asr\\data'
for name in listdir(dataset_path):
if isdir(join(dataset_path, name)):
print(name)
# Create an all targets list
all_targets = [name for name in listdir(dataset_path) if isdir(join(dataset_path, name))]
print(all_targets)
# Leave off background noise set
all_targets.remove('_background_noise_')
print(all_targets)
# See how many files are in each
num_samples = 0
for target in all_targets:
print(len(listdir(join(dataset_path, target))))
num_samples += len(listdir(join(dataset_path, target)))
print('Total samples:', num_samples)
# Settings
target_list = all_targets
feature_sets_file = 'all_targets_mfcc_sets.npz'
perc_keep_samples = 1.0 # 1.0 is keep all samples
val_ratio = 0.1
test_ratio = 0.1
sample_rate = 8000 # .wev file sampled in 16kHz
num_mfcc = 16
len_mfcc = 16
# Create list of filenames along with ground truth vector (y)
filenames = []
y = []
for index, target in enumerate(target_list):
print(join(dataset_path, target))
filenames.append(listdir(join(dataset_path, target)))
y.append(np.ones(len(filenames[index])) * index)
# Check ground truth Y vector
print(y)
for item in y:
print(len(item))
# Flatten filename and y vectors
filenames = [item for sublist in filenames for item in sublist]
y = [item for sublist in y for item in sublist]
# Associate filenames with true output and shuffle
filenames_y = list(zip(filenames, y))
random.shuffle(filenames_y)
filenames, y = zip(*filenames_y)
# Only keep the specified number of samples (shorter extraction/training)
print(len(filenames))
filenames = filenames[:int(len(filenames) * perc_keep_samples)]
print(len(filenames))
# Calculate validation and test set sizes
val_set_size = int(len(filenames) * val_ratio)
test_set_size = int(len(filenames) * test_ratio)
# Break dataset apart into train, validation, and test sets
filenames_val = filenames[:val_set_size]
filenames_test = filenames[val_set_size:(val_set_size + test_set_size)]
filenames_train = filenames[(val_set_size + test_set_size):]
# Break y apart into train, validation, and test sets
y_orig_val = y[:val_set_size]
y_orig_test = y[val_set_size:(val_set_size + test_set_size)]
y_orig_train = y[(val_set_size + test_set_size):]
# Function: Create MFCC from given path
def calc_mfcc(path):
# Load wavefile
signal, fs = librosa.load(path, sr=sample_rate)
# Create MFCCs from sound clip
mfccs = python_speech_features.base.mfcc(signal,
samplerate=fs,
winlen=0.256,
winstep=0.050,
numcep=num_mfcc,
nfilt=26,
nfft=2048,
preemph=0.0,
ceplifter=0,
appendEnergy=False,
winfunc=np.hanning)
return mfccs.transpose()
# TEST: Construct test set by computing MFCC of each WAV file
prob_cnt = 0
x_test = []
y_test = []
for index, filename in enumerate(filenames_train):
# Stop after 500
if index >= 500:
break
# Create path from given filename and target item
path = join(dataset_path, target_list[int(y_orig_train[index])],
filename)
# Create MFCCs
mfccs = calc_mfcc(path)
if mfccs.shape[1] == len_mfcc:
x_test.append(mfccs)
y_test.append(y_orig_train[index])
else:
print('Dropped:', index, mfccs.shape)
prob_cnt += 1
print('% of problematic samples:', prob_cnt / 500)
# !pip install playsound
# +
# TEST: Test shorter MFCC
from playsound import playsound
idx = 13
# Create path from given filename and target item
path = join(dataset_path, target_list[int(y_orig_train[idx])],
filenames_train[idx])
# Create MFCCs
mfccs = calc_mfcc(path)
print("MFCCs:", mfccs)
# Plot MFCC
fig = plt.figure()
plt.imshow(mfccs, cmap='inferno', origin='lower')
# TEST: Play problem sounds
print(target_list[int(y_orig_train[idx])])
playsound(path)
# -
# Function: Create MFCCs, keeping only ones of desired length
def extract_features(in_files, in_y):
prob_cnt = 0
out_x = []
out_y = []
for index, filename in enumerate(in_files):
# Create path from given filename and target item
path = join(dataset_path, target_list[int(in_y[index])],
filename)
# Check to make sure we're reading a .wav file
if not path.endswith('.wav'):
continue
# Create MFCCs
mfccs = calc_mfcc(path)
# Only keep MFCCs with given length
if mfccs.shape[1] == len_mfcc:
out_x.append(mfccs)
out_y.append(in_y[index])
else:
print('Dropped:', index, mfccs.shape)
prob_cnt += 1
return out_x, out_y, prob_cnt
# Create train, validation, and test sets
x_train, y_train, prob = extract_features(filenames_train,
y_orig_train)
print('Removed percentage:', prob / len(y_orig_train))
x_val, y_val, prob = extract_features(filenames_val, y_orig_val)
print('Removed percentage:', prob / len(y_orig_val))
x_test, y_test, prob = extract_features(filenames_test, y_orig_test)
print('Removed percentage:', prob / len(y_orig_test))
# Save features and truth vector (y) sets to disk
np.savez(feature_sets_file,
x_train=x_train,
y_train=y_train,
x_val=x_val,
y_val=y_val,
x_test=x_test,
y_test=y_test)
# TEST: Load features
feature_sets = np.load(feature_sets_file)
feature_sets.files
len(feature_sets['x_train'])
print(feature_sets['y_val'])
| 6,406 |
/Mofan/CNN_tensorflow.ipynb | ca85ad35f9f7dcd664f46dd713136960981886f0 | [] | no_license | luluenen/Moocs | https://github.com/luluenen/Moocs | 0 | 0 | null | null | null | null | Jupyter Notebook | false | false | .py | 4,633 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# +
from __future__ import print_function
import tensorflow as tf
from tensorflow.examples.tutorials.mnist import input_data
# number 1 to 10 data
mnist = input_data.read_data_sets('MNIST_data', one_hot=True)
def compute_accuracy(v_xs, v_ys):
global prediction
y_pre = sess.run(prediction, feed_dict={xs: v_xs, keep_prob: 1})
correct_prediction = tf.equal(tf.argmax(y_pre,1), tf.argmax(v_ys,1))
accuracy = tf.reduce_mean(tf.cast(correct_prediction, tf.float32))
result = sess.run(accuracy, feed_dict={xs: v_xs, ys: v_ys, keep_prob: 1})
return result
def weight_variable(shape):
initial = tf.truncated_normal(shape, stddev=0.1)
return tf.Variable(initial)
def bias_variable(shape):
initial = tf.constant(0.1, shape=shape)
return tf.Variable(initial)
def conv2d(x, W):
# stride [1, x_movement, y_movement, 1]
# Must have strides[0] = strides[3] = 1
return tf.nn.conv2d(x, W, strides=[1, 1, 1, 1], padding='SAME')
def max_pool_2x2(x):
# stride [1, x_movement, y_movement, 1]
return tf.nn.max_pool(x, ksize=[1,2,2,1], strides=[1,2,2,1], padding='SAME')
# define placeholder for inputs to network
xs = tf.placeholder(tf.float32, [None, 784])/255. # 28x28
ys = tf.placeholder(tf.float32, [None, 10])
keep_prob = tf.placeholder(tf.float32)
x_image = tf.reshape(xs, [-1, 28, 28, 1])
# print(x_image.shape) # [n_samples, 28,28,1]
## conv1 layer ##
W_conv1 = weight_variable([5,5, 1,32]) # patch 5x5, in size 1, out size 32
b_conv1 = bias_variable([32])
h_conv1 = tf.nn.relu(conv2d(x_image, W_conv1) + b_conv1) # output size 28x28x32
h_pool1 = max_pool_2x2(h_conv1) # output size 14x14x32
## conv2 layer ##
W_conv2 = weight_variable([5,5, 32, 64]) # patch 5x5, in size 32, out size 64
b_conv2 = bias_variable([64])
h_conv2 = tf.nn.relu(conv2d(h_pool1, W_conv2) + b_conv2) # output size 14x14x64
h_pool2 = max_pool_2x2(h_conv2) # output size 7x7x64
## fc1 layer ##
W_fc1 = weight_variable([7*7*64, 1024])
b_fc1 = bias_variable([1024])
# [n_samples, 7, 7, 64] ->> [n_samples, 7*7*64]
h_pool2_flat = tf.reshape(h_pool2, [-1, 7*7*64])
h_fc1 = tf.nn.relu(tf.matmul(h_pool2_flat, W_fc1) + b_fc1)
h_fc1_drop = tf.nn.dropout(h_fc1, keep_prob)
## fc2 layer ##
W_fc2 = weight_variable([1024, 10])
b_fc2 = bias_variable([10])
prediction = tf.nn.softmax(tf.matmul(h_fc1_drop, W_fc2) + b_fc2)
# the error between prediction and real data
cross_entropy = tf.reduce_mean(-tf.reduce_sum(ys * tf.log(prediction),
reduction_indices=[1])) # loss
train_step = tf.train.AdamOptimizer(1e-4).minimize(cross_entropy)
sess = tf.Session()
# important step
# tf.initialize_all_variables() no long valid from
# 2017-03-02 if using tensorflow >= 0.12
if int((tf.__version__).split('.')[1]) < 12 and int((tf.__version__).split('.')[0]) < 1:
init = tf.initialize_all_variables()
else:
init = tf.global_variables_initializer()
sess.run(init)
for i in range(1000):
batch_xs, batch_ys = mnist.train.next_batch(100)
sess.run(train_step, feed_dict={xs: batch_xs, ys: batch_ys, keep_prob: 0.5})
if i % 50 == 0:
print(compute_accuracy(
mnist.test.images[:1000], mnist.test.labels[:1000]))
| 3,556 |
/the_data_science_process/Putting It All Together.ipynb | 111aa1effffa96817610d150b558964e006afcc3 | [] | no_license | lewi0332/udacity-data-science-nanodegree | https://github.com/lewi0332/udacity-data-science-nanodegree | 0 | 0 | null | null | null | null | Jupyter Notebook | false | false | .py | 86,733 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# #### Putting It All Together
#
# As you might have guessed from the last notebook, using all of the variables was allowing you to drastically overfit the training data. This was great for looking good in terms of your Rsquared on these points. However, this was not great in terms of how well you were able to predict on the test data.
#
# We will start where we left off in the last notebook. First read in the dataset.
# +
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from sklearn.linear_model import LinearRegression
from sklearn.model_selection import train_test_split
from sklearn.metrics import r2_score, mean_squared_error
import AllTogether as t
import seaborn as sns
# %matplotlib inline
df = pd.read_csv('./survey_results_public.csv')
df.head()
# -
# #### Question 1
#
# **1.** To begin fill in the format function below with the correct variable. Notice each **{ }** holds a space where one of your variables will be added to the string. This will give you something to do while the the function does all the steps you did throughout this lesson.
# +
a = 'test_score'
b = 'train_score'
c = 'linear model (lm_model)'
d = 'X_train and y_train'
e = 'X_test'
f = 'y_test'
g = 'train and test data sets'
h = 'overfitting'
q1_piat = '''In order to understand how well our {} fit the dataset,
we first needed to split our data into {}.
Then we were able to fit our {} on the {}.
We could then predict using our {} by providing
the linear model the {} for it to make predictions.
These predictions were for {}.
By looking at the {}, it looked like we were doing awesome because
it was 1! However, looking at the {} suggested our model was not
extending well. The purpose of this notebook will be to see how
well we can get our model to extend to new data.
This problem where our data fits the training data well, but does
not perform well on test data is commonly known as
{}.'''.format(c, g, c, d, c, e, f, b, a, h) #replace a with the correct variable
print(q1_piat)
# -
# Print the solution order of the letters in the format
t.q1_piat_answer()
# #### Question 2
#
# **2.** Now, we need to improve the model . Use the dictionary below to provide the true statements about improving **this model**. **Also consider each statement as a stand alone**. Though, it might be a good idea after other steps, which would you consider a useful **next step**?
# +
a = 'yes'
b = 'no'
q2_piat = {'add interactions, quadratics, cubics, and other higher order terms': b,
'fit the model many times with different rows, then average the responses': a,
'subset the features used for fitting the model each time': a,
'this model is hopeless, we should start over': b}
# -
#Check your solution
t.q2_piat_check(q2_piat)
# ##### Question 3
#
# **3.** Before we get too far along, follow the steps in the function below to create the X (explanatory matrix) and y (response vector) to be used in the model. If your solution is correct, you should see a plot similar to the one shown in the Screencast.
# +
def clean_data(df):
'''
INPUT
df - pandas dataframe
OUTPUT
X - A matrix holding all of the variables you want to consider when predicting the response
y - the corresponding response vector
Perform to obtain the correct X and y objects
This function cleans df using the following steps to produce X and y:
1. Drop all the rows with no salaries
2. Create X as all the columns that are not the Salary column
3. Create y as the Salary column
4. Drop the Salary, Respondent, and the ExpectedSalary columns from X
5. For each numeric variable in X, fill the column with the mean value of the column.
6. Create dummy columns for all the categorical variables in X, drop the original columns
'''
df.dropna(subset=['Salary'], axis=0, inplace=True)
df.drop(['Respondent', 'ExpectedSalary'], axis=1, inplace=True)
df.dropna(axis=1, how='all', inplace=True)
df = df.apply(lambda col: col.fillna(col.mean()) if col.dtype != 'object' else col)
cat_cols = df.select_dtypes(include='object').columns
df = df.drop(cat_cols, axis=1).join(pd.get_dummies(df[cat_cols], prefix=cat_cols, dummy_na=False))
y = df.pop('Salary')
return df, y
#Use the function to create X and y
X, y = clean_data(df)
# -
# ### Run the Cell Below to Acheive the Results Needed for Question 4
# +
#cutoffs here pertains to the number of missing values allowed in the used columns.
#Therefore, lower values for the cutoff provides more predictors in the model.
cutoffs = [5000, 3500, 2500, 1000, 100, 50, 30, 25]
#Run this cell to pass your X and y to the model for testing
r2_scores_test, r2_scores_train, lm_model, X_train, X_test, y_train, y_test = t.find_optimal_lm_mod(X, y, cutoffs)
# -
# #### Question 4
#
# **4.** Use the output and above plot to correctly fill in the keys of the **q4_piat** dictionary with the correct variable. Notice that only the optimal model results are given back in the above - they are stored in **lm_model**, **X_train**, **X_test**, **y_train**, and **y_test**. If more than one answer holds, provide a tuple holding all the correct variables in the order of first variable alphabetically to last variable alphabetically.
# Cell for your computations to answer the next question
r2_scores_train
# +
a = 'we would likely have a better rsquared for the test data.'
b = 1000
c = 872
d = 0.69
e = 0.82
f = 0.88
g = 0.72
h = 'we would likely have a better rsquared for the training data.'
q4_piat = {'The optimal number of features based on the results is': c,
'The model we should implement in practice has a train rsquared of': e,
'The model we should implement in practice has a test rsquared of': d,
'If we were to allow the number of features to continue to increase':h
}
# -
#Check against your solution
t.q4_piat_check(q4_piat)
# #### Question 5
#
# **5.** The default penalty on coefficients using linear regression in sklearn is a ridge (also known as an L2) penalty. Because of this penalty, and that all the variables were normalized, we can look at the size of the coefficients in the model as an indication of the impact of each variable on the salary. The larger the coefficient, the larger the expected impact on salary.
#
# Use the space below to take a look at the coefficients. Then use the results to provide the **True** or **False** statements based on the data.
#
# #### Run the below to complete the following dictionary
coefs_df = coefs_df.sort_values('abs_coefs', ascending=False)
# +
def coef_weights(coefficients, X_train):
'''
INPUT:
coefficients - the coefficients of the linear model
X_train - the training data, so the column names can be used
OUTPUT:
coefs_df - a dataframe holding the coefficient, estimate, and abs(estimate)
Provides a dataframe that can be used to understand the most influential coefficients
in a linear model by providing the coefficient estimates along with the name of the
variable attached to the coefficient.
'''
coefs_df = pd.DataFrame()
coefs_df['est_int'] = X_train.columns
coefs_df['coefs'] = lm_model.coef_
coefs_df['abs_coefs'] = np.abs(lm_model.coef_)
coefs_df = coefs_df.sort_values('abs_coefs', ascending=False)
coefs_df['abs_coefs'] = coefs_df['abs_coefs'].astype('int')
return coefs_df
#Use the function
coef_df = coef_weights(lm_model.coef_, X_train)
#A quick look at the top results
coef_df.head(20)
# +
a = True
b = False
#According to the data...
q5_piat = {'Country appears to be one of the top indicators for salary': a,
'Gender appears to be one of the indicators for salary': b,
'How long an individual has been programming appears to be one of the top indicators for salary': a,
'The longer an individual has been programming the more they are likely to earn': b}
# -
t.q5_piat_check(q5_piat)
# #### Congrats of some kind
#
# Congrats! Hopefully this was a great review, or an eye opening experience about how to put the steps together for an analysis. List the steps. In the next lesson, you will look at how take this and show it off to others so they can act on it.
| 8,757 |
/day_17/day17.ipynb | e4befcd43eab6067a026d2970b5cf30a1dd60820 | [] | no_license | mataln/aoc_julia_2020 | https://github.com/mataln/aoc_julia_2020 | 0 | 0 | null | null | null | null | Jupyter Notebook | false | false | .jl | 4,401 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .jl
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Julia 1.5.3
# language: julia
# name: julia-1.5
# ---
example_input = open(f->read(f, String), "example.txt")
task_input = open(f->read(f, String), "task.txt")
# +
using Images
function pad_grid(grid::Array{Char,3})
sz = size(grid)[1]
new_grid = fill('.', (sz+2,sz+2,sz+2))
new_grid[2:end-1,2:end-1,2:end-1] = grid
return new_grid
end
function count_neighbours(coords::Array{Int64,1}, grid::Array{Char,3})
sz = size(grid)[1]
x, y, z = coords[1], coords[2], coords[3]
adjacent_count = 0
for x2 in max(x-1,1):min(x+1,sz)
for y2 in max(y-1,1):min(y+1,sz)
for z2 in max(z-1,1):min(z+1,sz)
(!([x2,y2,z2] == [x,y,z]) && grid[x2,y2,z2] == '#') && (adjacent_count += 1)
end
end
end
return adjacent_count
end
function task1(puzzle_input::String)
rows = split(puzzle_input, "\n")
sz = length(rows)
grid = fill('.', (sz, sz, sz))
for (i,row) in enumerate(rows)
grid[i,:,1] = [x for x in row]
end
grid = pad_grid(grid)
for _ in 1:6
sz = size(grid)[1]
new_grid = fill('.', (sz, sz, sz))
for x in 1:sz
for y in 1:sz
for z in 1:sz
neighbours = count_neighbours([x,y,z], grid)
if grid[x, y, z] == '#'
(neighbours == 2 || neighbours == 3) && (new_grid[x, y, z] = '#')
else
neighbours == 3 && (new_grid[x, y, z] = '#')
end
end
end
end
grid = pad_grid(new_grid)
end
return count(i->i=='#', grid)
end
# -
grid = task1(task_input)
count_neighbours([2,4,2], grid)
| 1,994 |
/demos/community_detection/attacks_clustering_analysis.ipynb | 9b31b6f569f59950f5cbfcd5e10668fd9996896f | [
"Apache-2.0"
] | permissive | rpatil524/stellargraph | https://github.com/rpatil524/stellargraph | 0 | 0 | Apache-2.0 | 2021-10-29T21:37:14 | 2021-10-11T22:28:18 | Python | Jupyter Notebook | false | false | .py | 668,822 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Comparison of clustering of node embeddings with a traditional community detection method
#
# + [markdown] nbsphinx="hidden" tags=["CloudRunner"]
# <table><tr><td>Run the latest release of this notebook:</td><td><a href="https://mybinder.org/v2/gh/stellargraph/stellargraph/master?urlpath=lab/tree/demos/community_detection/attacks_clustering_analysis.ipynb" alt="Open In Binder" target="_parent"><img src="https://mybinder.org/badge_logo.svg"/></a></td><td><a href="https://colab.research.google.com/github/stellargraph/stellargraph/blob/master/demos/community_detection/attacks_clustering_analysis.ipynb" alt="Open In Colab" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg"/></a></td></tr></table>
# -
# ## Introduction
# The goal of this use case is to demonstrate how node embeddings from graph convolutional neural networks trained in unsupervised manner are comparable to standard community detection methods based on graph partitioning.
#
# Specifically, we use unsupervised [graphSAGE](http://snap.stanford.edu/graphsage/) approach to learn node embeddings of terrorist groups in a publicly available dataset of global terrorism events, and analyse clusters of these embeddings. We compare the clusters to communities produced by [Infomap](http://www.mapequation.org), a state-of-the-art approach of graph partitioning based on information theory.
#
# We argue that clustering based on unsupervised graphSAGE node embeddings allow for richer representation of the data than its graph partitioning counterpart as the former takes into account node features together with the graph structure, while the latter utilises only the graph structure.
#
# We demonstrate, using the terrorist group dataset, that the infomap communities and the graphSAGE embedding clusters (GSEC) provide qualitatively different insights into underlying data patterns.
# ### Data description
# __The Global Terrorism Database (GTD)__ used in this demo is available here: https://www.kaggle.com/START-UMD/gtd. GTD is an open-source database including information on terrorist attacks around the world from 1970 through 2017. The GTD includes systematic data on domestic as well as international terrorist incidents that have occurred during this time period and now includes more than 180,000 attacks. The database is maintained by researchers at the National Consortium for the Study of Terrorism and Responses to Terrorism (START), headquartered at the University of Maryland. For information refer to the initial data source: https://www.start.umd.edu/gtd/.
#
# Full dataset contains information on more than 180,000 Terrorist Attacks.
# ### Glossary:
# For this particular study we adopt the following terminology:
#
# - __a community__ is a group of nodes produced by a traditional community detection algorithm (infomap community in this use case)
# - __a cluster__ is a group of nodes that were clustered together using a clustering algorithm applied to node embeddings (here, [DBSCAN clustering](https://www.aaai.org/Papers/KDD/1996/KDD96-037.pdf) applied to unsupervised GraphSAGE embeddings).
#
# For more detailed explanation of unsupervised graphSAGE see [Unsupervised graphSAGE demo](../embeddings/graphsage-unsupervised-sampler-embeddings.ipynb).
# The rest of the demo is structured as follows. First, we load the data and preprocess it (see `utils.py` for detailed steps of network and features generation). Then we apply infomap and visualise results of one selected community obtained from this method. Next, we apply unsupervised graphSAGE on the same data and extract node embeddings. We first tune DBSCAN hyperparameters, and apply DBSCAN that produces sufficient numbers of clusters with minimal number of noise points. We look at the resulting clusters, and investigate a single selected cluster in terms of the graph structure and features of the nodes in this cluster. Finally, we conclude our investigation with the summary of the results.
# + nbsphinx="hidden" tags=["CloudRunner"]
# install StellarGraph if running on Google Colab
import sys
if 'google.colab' in sys.modules:
# %pip install -q stellargraph[demos]==1.3.0b
# + nbsphinx="hidden" tags=["VersionCheck"]
# verify that we're using the correct version of StellarGraph for this notebook
import stellargraph as sg
try:
sg.utils.validate_notebook_version("1.3.0b")
except AttributeError:
raise ValueError(
f"This notebook requires StellarGraph version 1.3.0b, but a different version {sg.__version__} is installed. Please see <https://github.com/stellargraph/stellargraph/issues/1172>."
) from None
# +
import pandas as pd
import numpy as np
import networkx as nx
import igraph as ig
from sklearn.cluster import DBSCAN
from sklearn import metrics
from sklearn import preprocessing, feature_extraction, model_selection
from sklearn.decomposition import PCA
from sklearn.manifold import TSNE
import random
import matplotlib.pyplot as plt
from matplotlib.pyplot import figure
# %matplotlib inline
import warnings
warnings.filterwarnings('ignore') # supress warnings due to some future deprications
# +
import stellargraph as sg
from stellargraph.data import EdgeSplitter
from stellargraph.mapper import GraphSAGELinkGenerator, GraphSAGENodeGenerator
from stellargraph.layer import GraphSAGE, link_classification
from stellargraph.data import UniformRandomWalk
from stellargraph.data import UnsupervisedSampler
from sklearn.model_selection import train_test_split
from tensorflow import keras
from stellargraph import globalvar
# -
import mplleaflet
from itertools import count
import utils
# ### Data loading
# This is the raw data of terrorist attacks that we use as a starting point of our analysis.
dt_raw = pd.read_csv(
"~/data/globalterrorismdb_0718dist.csv",
sep=",",
engine="python",
encoding="ISO-8859-1",
)
# ### Loading preprocessed features
# The resulting feature set is the aggregation of features for each of the terrorist groups (`gname`). We collect such features as total number of attacks per terrorist group, number of perpetrators etc. We use `targettype` and `attacktype` (number of attacks/targets of particular type) and transform it to a wide representation of data (e.g each type is a separate column with the counts for each group). Refer to `utils.py` for more detailed steps of the preprocessing and data cleaning.
gnames_features = utils.load_features(input_data=dt_raw)
gnames_features.head()
# ### Edgelist creation
# The raw dataset contains information for the terrotist attacks. It contains information about some incidents being related. However, the resulting graph is very sparse. Therefore, we create our own schema for the graph, where two terrorist groups are connected, if they had at least one terrorist attack in the same country and in the same decade. To this end we proceed as follows:
#
# - we group the event data by `gname` - the names of the terrorist organisations.
# - we create a new feature `decade` based on `iyear`, where one bin consists of 10 years (attacks of 70s, 80s, and so on).
# - we add the concatination of of the decade and the country of the attack: `country_decade` which will become a link between two terrorist groups.
# - finally, we create an edgelist, where two terrorist groups are linked if they have operated in the same country in the same decade. Edges are undirected.
# In addition, some edges are created based on the column `related` in the raw event data. The description of the data does not go into details how these incidents were related. We utilise this information creating a link for terrorist groups if the terrorist attacks performed by these groups were related. If related events corresponded to the same terrorist group, they were discarded (we don't use self-loops in the graph). However, the majority of such links are already covered by `country_decade` edge type. Refer to `utils.py` for more information on graph creation.
G = utils.load_network(input_data=dt_raw)
print(nx.info(G))
# ### Connected components of the network
# Note that the graph is disconnected, consisting of 21 connected components.
print(nx.number_connected_components(G))
# Get the sizes of the connected components:
Gcc = sorted(nx.connected_component_subgraphs(G), key=len, reverse=True)
cc_sizes = []
for cc in list(Gcc):
cc_sizes.append(len(cc.nodes()))
print(cc_sizes)
# The distribution of connected components' sizes shows that there is a single large component, and a few isolated groups. We expect the community detection/node embedding clustering algorithms discussed below to discover non-trivial communities that are not simply the connected components of the graph.
# ## Traditional community detection
# We perform traditional community detection via `infomap` implemented in `igraph`. We translate the original `networkx` graph object to `igraph`, apply `infomap` to it to detect communities, and assign the resulting community memberships back to the `networkx` graph.
# translate the object into igraph
g_ig = ig.Graph.Adjacency(
(nx.to_numpy_matrix(G) > 0).tolist(), mode=ig.ADJ_UNDIRECTED
) # convert via adjacency matrix
g_ig.summary()
# perform community detection
random.seed(123)
c_infomap = g_ig.community_infomap()
print(c_infomap.summary())
# We get 160 communities, meaning that the largest connected components of the graph are partitioned into more granular groups.
# plot the community sizes
infomap_sizes = c_infomap.sizes()
plt.title("Infomap community sizes")
plt.xlabel("community id")
plt.ylabel("number of nodes")
plt.bar(list(range(1, len(infomap_sizes) + 1)), infomap_sizes)
# Modularity metric for infomap
c_infomap.modularity
# The discovered infomap communities have smooth distribution of cluster sizes, which indicates that the underlying graph structure has a natural partitioning. The modularity score is also pretty high indicating that nodes are more tightly connected within clusters than expected from random graph, i.e., the discovered communities are tightly-knit.
# assign community membership results back to networkx, keep the dictionary for later comparisons with the clustering
infomap_com_dict = dict(zip(list(G.nodes()), c_infomap.membership))
nx.set_node_attributes(G, infomap_com_dict, "c_infomap")
# ### Visualisation of the infomap communities
# We can visualise the resulting communities using maps as the constructed network is based partially on the geolocation. The raw data have a lat-lon coordinates for each of the attacks. Terrorist groups might perform attacks in different locations. However, currently the simplified assumption is made, and the average of latitude and longitude for each terrorist group is calculated. Note that it might result in some positions being "off", but it is good enough to provide a glimpse whether the communities are consistent with the location of that groups.
# +
# fill NA based on the country name
dt_raw.latitude[
dt_raw["gname"] == "19th of July Christian Resistance Brigade"
] = 12.136389
dt_raw.longitude[
dt_raw["gname"] == "19th of July Christian Resistance Brigade"
] = -86.251389
# filter only groups that are present in a graph
dt_filtered = dt_raw[dt_raw["gname"].isin(list(G.nodes()))]
# collect averages of latitude and longitude for each of gnames
avg_coords = dt_filtered.groupby("gname")[["latitude", "longitude"]].mean()
print(avg_coords.shape)
print(len(G.nodes()))
# -
# As plotting the whole graph is not feasible, we investigate a single community
# __Specify community id in the range of infomap total number of clusters__ (`len(infomap_sizes)`)
com_id = 50 # smaller number - larger community, as it's sorted
# extraction of a subgraph from the nodes in this community
com_G = G.subgraph(
[n for n, attrdict in G.nodes.items() if attrdict["c_infomap"] == com_id]
)
print(nx.info(com_G))
# plot community structure only
pos = nx.random_layout(com_G, seed=123)
plt.figure(figsize=(10, 8))
nx.draw_networkx(com_G, pos, edge_color="#26282b", node_color="blue", alpha=0.3)
plt.axis("off")
plt.show()
# +
# plot on the map
nodes = com_G.nodes()
com_avg_coords = avg_coords[avg_coords.index.isin(list(nodes))]
com_avg_coords.fillna(
com_avg_coords.mean(), inplace=True
) # fill missing values with the average
new_order = [1, 0]
com_avg_coords = com_avg_coords[com_avg_coords.columns[new_order]]
pos = com_avg_coords.T.to_dict("list") # layout is based on the provided coordindates
fig, ax = plt.subplots(figsize=(12, 6))
nx.draw_networkx_edges(com_G, pos, edge_color="grey")
nx.draw_networkx_nodes(com_G, pos, nodelist=nodes, node_size=200, alpha=0.5)
nx.draw_networkx_labels(com_G, pos, font_color="#362626", font_size=50)
mplleaflet.display(fig=ax.figure)
# -
# (**N.B.:** the above interactive plot will only appear after running the cell, and is not rendered in GitHub!)
# ### Summary of results based on infomap
# Infomap is a robust community detection algorithm, and shows good results that are in line with the expectations. Most of the communities are tightly connected and reflect the geographical position of the events of the terrorist groups. That is because the graph schema is expressed as
# > _two terrorist groups are connected if they have terrorist events in the same country in the same decade_.
#
# However, no node features are taken into account in the case of the traditional community detection.
#
# Next, we explore the GSEC approach, where node features are used along with the graph structure.
# ## Node represenatation learning with unsupervised graphSAGE
# Now we apply unsupervised GraphSAGE that takes into account node features as well as graph structure, to produce *node embeddings*. In our case, similarity of node embeddings depicts similarity of the terrorist groups in terms of their operations, targets and attack types (node features) as well as in terms of time and place of attacks (graph structure).
# +
# we reload the graph to get rid of assigned attributes
G = utils.load_network(input_data=dt_raw) # to make sure that graph is clean
# filter features to contain only gnames that are among nodes of the network
filtered_features = gnames_features[gnames_features["gname"].isin(list(G.nodes()))]
filtered_features.set_index("gname", inplace=True)
filtered_features.shape
# -
filtered_features.head() # take a glimpse at the feature data
# We perform a log-transform of the feature set to rescale feature values.
# transforming features to be on log scale
node_features = filtered_features.transform(lambda x: np.log1p(x))
# sanity check that there are no misspelled gnames left
set(list(G.nodes())) - set(list(node_features.index.values))
# ### Unsupervised graphSAGE
#
# (For a detailed unsupervised GraphSAGE workflow with a narrative, see [Unsupervised graphSAGE demo](../embeddings/graphsage-unsupervised-sampler-embeddings.ipynb))
Gs = sg.StellarGraph.from_networkx(G, node_features=node_features)
print(Gs.info())
# parameter specification
number_of_walks = 3
length = 5
batch_size = 50
epochs = 10
num_samples = [20, 20]
layer_sizes = [100, 100]
learning_rate = 1e-2
unsupervisedSamples = UnsupervisedSampler(
Gs, nodes=G.nodes(), length=length, number_of_walks=number_of_walks
)
generator = GraphSAGELinkGenerator(Gs, batch_size, num_samples)
train_gen = generator.flow(unsupervisedSamples)
# +
assert len(layer_sizes) == len(num_samples)
graphsage = GraphSAGE(
layer_sizes=layer_sizes, generator=generator, bias=True, dropout=0.0, normalize="l2"
)
# -
# We now build a Keras model from the GraphSAGE class that we can use for unsupervised predictions. We add a `link_classfication` layer as unsupervised training operates on node pairs.
# +
x_inp, x_out = graphsage.in_out_tensors()
prediction = link_classification(
output_dim=1, output_act="sigmoid", edge_embedding_method="ip"
)(x_out)
# +
model = keras.Model(inputs=x_inp, outputs=prediction)
model.compile(
optimizer=keras.optimizers.Adam(lr=learning_rate),
loss=keras.losses.binary_crossentropy,
metrics=[keras.metrics.binary_accuracy],
)
# -
history = model.fit(
train_gen,
epochs=epochs,
verbose=2,
use_multiprocessing=False,
workers=1,
shuffle=True,
)
# ### Extracting node embeddings
node_ids = list(Gs.nodes())
node_gen = GraphSAGENodeGenerator(Gs, batch_size, num_samples).flow(node_ids)
embedding_model = keras.Model(inputs=x_inp[::2], outputs=x_out[0])
node_embeddings = embedding_model.predict(node_gen, workers=4, verbose=1)
# #### 2D t-sne plot of the resulting node embeddings
# Here we visually check whether embeddings have some underlying cluster structure.
node_embeddings.shape
# +
# TSNE visualisation to check whether the embeddings have some structure:
X = node_embeddings
if X.shape[1] > 2:
transform = TSNE # PCA
trans = transform(n_components=2, random_state=123)
emb_transformed = pd.DataFrame(trans.fit_transform(X), index=node_ids)
else:
emb_transformed = pd.DataFrame(X, index=node_ids)
emb_transformed = emb_transformed.rename(columns={"0": 0, "1": 1})
alpha = 0.7
fig, ax = plt.subplots(figsize=(7, 7))
ax.scatter(emb_transformed[0], emb_transformed[1], alpha=alpha)
ax.set(aspect="equal", xlabel="$X_1$", ylabel="$X_2$")
plt.title("{} visualization of GraphSAGE embeddings".format(transform.__name__))
plt.show()
# -
# #### t-sne colored by infomap
# We also depict the same t-sne plot colored by infomap communities. As we can observe t-sne of GraphSAGE embeddings do not really separate the infomap communities.
emb_transformed["infomap_clusters"] = emb_transformed.index.map(infomap_com_dict)
plt.scatter(
emb_transformed[0],
emb_transformed[1],
c=emb_transformed["infomap_clusters"],
cmap="Spectral",
edgecolors="black",
alpha=0.3,
s=100,
)
plt.title("t-sne with colors corresponding to infomap communities")
# Next, we apply dbscan algorithm to cluster the embeddings. dbscan has two hyperparameters: `eps` and `min_samples`, and produces clusters along with the noise points (the points that could not be assigned to any particular cluster, indicated as -1). These tunable parameters directly affect the cluster results. We use greedy search over the hyperparameters and check what are the good candidates.
db_dt = utils.dbscan_hyperparameters(
node_embeddings, e_lower=0.1, e_upper=0.9, m_lower=5, m_upper=15
)
# print results where there are more clusters than 1, and sort by the number of noise points
db_dt.sort_values(by=["n_noise"])[db_dt.n_clusters > 1]
# Pick the hyperparameters, where the clustering results have as little noise points as possible, but also create number of clusters of reasonable size.
# perform dbscan with the chosen parameters:
db = DBSCAN(eps=0.1, min_samples=5).fit(node_embeddings)
# Calculating the clustering statistics:
labels = db.labels_
# Number of clusters in labels, ignoring noise if present.
n_clusters_ = len(set(labels)) - (1 if -1 in labels else 0)
n_noise_ = list(labels).count(-1)
print("Estimated number of clusters: %d" % n_clusters_)
print("Estimated number of noise points: %d" % n_noise_)
print("Silhouette Coefficient: %0.3f" % metrics.silhouette_score(node_embeddings, labels))
# We plot t-sne again but with the colours corresponding to dbscan points.
# +
emb_transformed["dbacan_clusters"] = labels
X = emb_transformed[emb_transformed["dbacan_clusters"] != -1]
plt.scatter(
X[0],
X[1],
c=X["dbacan_clusters"],
cmap="Spectral",
edgecolors="black",
alpha=0.3,
s=100,
)
plt.title("t-sne with colors corresponding to dbscan cluster. Without noise points")
# -
# ### Investigating GSEC and infomap qualitative differences
# Let's take a look at the resulting GSEC clusters, and explore, as an example, one particular cluster of a reasonable size, which is not a subset of any single infomap community.
# Display cluster sizes for first 15 largest clusters:
clustered_df = pd.DataFrame(node_embeddings, index=node_ids)
clustered_df["cluster"] = db.labels_
clustered_df.groupby("cluster").count()[0].sort_values(ascending=False)[0:15]
# We want to display clusters that differ from infomap communities, as they are more interesting in this context. Therefore we calculate for each DBSCAN cluster how many different infomap communities it contains. The results are displayed below.
inf_db_cm = clustered_df[["cluster"]]
inf_db_cm["infomap"] = inf_db_cm.index.map(infomap_com_dict)
dbscan_different = inf_db_cm.groupby("cluster")[
"infomap"
].nunique() # if 1 all belong to same infomap cluster
# show only those clusters that are not the same as infomap
dbscan_different[dbscan_different != 1]
# For example, DBSCAN `cluster_12` has nodes that were assigned to 2 different infomap clusters, while `cluster_31` has nodes from 8 different infomap communities.
# ### Single cluster visualisation
#
# Now that we've selected a GSEC cluster (id=20) of reasonable size that contains nodes belonging to 4 different infomap communities, let's explore it.
# __To visualise a particular cluster, specify its number here:__
# specify the cluster id here:
cluster_id = 20
# manually look at the terrorist group names
list(clustered_df.index[clustered_df.cluster == cluster_id])
# create a subgraph from the nodes in the cluster
cluster_G = G.subgraph(list(clustered_df.index[clustered_df.cluster == cluster_id]))
# List for each of the `gname` (terrorist group name) in the cluster the assigned infomap community id. This shows whether the similar community was produced by infomap or not.
comparison = {
k: v
for k, v in infomap_com_dict.items()
if k in list(clustered_df.index[clustered_df.cluster == cluster_id])
}
comparison
# As another metric of clustering quality, we display how many edges are inside this cluster vs how many nodes are going outside (only one of edge endpoints is inside a cluster)
external_internal_edges = utils.cluster_external_internal_edges(G, inf_db_cm, "cluster")
external_internal_edges[external_internal_edges.cluster == cluster_id]
# plot the structure only
pos = nx.fruchterman_reingold_layout(cluster_G, seed=123, iterations=30)
plt.figure(figsize=(10, 8))
nx.draw_networkx(cluster_G, pos, edge_color="#26282b", node_color="blue", alpha=0.3)
plt.axis("off")
plt.show()
# Recall that terrorist groups (nodes) are connected, when at least one of the attacks was performed in the same decade in the same country. Therefore the connectivity indicates spatio-temporal similarity.
# There are quite a few clusters that are similar to infomap clusters. We pick a cluster that is not a subset of any single infomap community. We can see that there are disconnected groups of nodes in this cluster.
#
# So why are these disconnected components combined into one cluster? GraphSAGE embeddings directly depend on both node features and an underlying graph structure. Therefore, it makes sense to investigate similarity of features of the nodes in the cluster. It can highlight why these terrorist groups are combined together by GSEC.
cluster_feats = filtered_features[
filtered_features.index.isin(
list(clustered_df.index[clustered_df.cluster == cluster_id])
)
]
# show only non-zero columns
features_nonzero = cluster_feats.loc[:, (cluster_feats != 0).any()]
features_nonzero.style.background_gradient(cmap="RdYlGn_r")
# We can see that most of the isolated nodes in the cluster have features similar to those in the tight clique, e.g., in most cases they have high number of attacks, high success ratio, and attacks being focused mostly on bombings, and their targets are often the police.
#
# Note that there are terrorist groups that differ from the rest of the groups in the cluster in terms of their features. By taking a closer look we can observe that these terrorist groups are a part of a tight clique. For example, _Martyr al-Nimr Battalion_ has number of bombings equal to 0, but it is a part of a fully connected subgraph.
#
# Interestingly, _Al-Qaida in Saudi Arabia_ ends up in the same cluster as _Al-Qaida in Yemen_, though they are not connected directly in the network.
#
# Thus we can observe that clustering on GraphSAGE embeddings combines groups based both on the underlying structure as well as features.
# +
nodes = cluster_G.nodes()
com_avg_coords = avg_coords[avg_coords.index.isin(list(nodes))]
new_order = [1, 0]
com_avg_coords = com_avg_coords[com_avg_coords.columns[new_order]]
com_avg_coords.fillna(
com_avg_coords.mean(), inplace=True
) # fill missing values with the average
pos = com_avg_coords.T.to_dict("list") # layout is based on the provided coordindates
fig, ax = plt.subplots(figsize=(22, 12))
nx.draw_networkx_nodes(cluster_G, pos, nodelist=nodes, node_size=200, alpha=0.5)
nx.draw_networkx_labels(cluster_G, pos, font_color="red", font_size=50)
nx.draw_networkx_edges(cluster_G, pos, edge_color="grey")
mplleaflet.display(fig=ax.figure)
# -
# (**N.B.:** the above interactive plot will only appear after running the cell, and is not rendered in GitHub!)
# These groups are also very closely located, but in contrast with infomap, this is not the only thing that defines the clustering groups.
# #### GSEC results
#
# What we can see from the results above is a somehow different picture from the infomap (note that by rerunning this notebook, the results might change due to the nature of the GraphSAGE predictions). Some clusters are identical to the infomap, showing that GSEC __capture__ the network structure. But some of the clusters differ - they are not necessary connected. By observing the feature table we can see that it has terrorist groups with __similar characteristics__.
# ## Conclusion
# In this use case we demonstrated the conceptual differences of traditional community detection and unsupervised GraphSAGE embeddings clustering (GSEC) on a real dataset. We can observe that the traditional approach to the community detection via graph partitioning produces communities that are related to the graph structure only, while GSEC combines the features and the structure. However, in the latter case we should be very conscious of the network schema and the features, as these significantly affect both the clustering and the interpretation of the resulting clusters.
#
# For example, if we don't want the clusters where neither number of kills nor country play a role, we might want to exclude `nkills` from the feature set, and create a graph, where groups are connected only via the decade, and not via country. Then, the resulting groups would probably be related to the terrorist activities in time, and grouped by their similarity in their target and the types of attacks.
# + [markdown] nbsphinx="hidden" tags=["CloudRunner"]
# <table><tr><td>Run the latest release of this notebook:</td><td><a href="https://mybinder.org/v2/gh/stellargraph/stellargraph/master?urlpath=lab/tree/demos/community_detection/attacks_clustering_analysis.ipynb" alt="Open In Binder" target="_parent"><img src="https://mybinder.org/badge_logo.svg"/></a></td><td><a href="https://colab.research.google.com/github/stellargraph/stellargraph/blob/master/demos/community_detection/attacks_clustering_analysis.ipynb" alt="Open In Colab" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg"/></a></td></tr></table>
| 27,683 |
/ro60_analysis.ipynb | bec2e8df2cd6436587a05987084b7881d774bf90 | [] | no_license | gpratt/tscc_ipython | https://github.com/gpratt/tscc_ipython | 0 | 0 | null | null | null | null | Jupyter Notebook | false | false | .py | 4,604,206 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Python 2
# language: python
# name: python2
# ---
from IPython.core.display import HTML
import pandas as pd
results = pd.read_table("/home/gpratt/projects/tiffany_genentech/analysis/ad-hoc/LIB19982_SAM18146099_R1_resequenced.R08.polyATrim.adapterTrim.Aligned.out.sorted.rmDup.sorted.0x100.peaks_annotated.txt")
foo = results.groupby('Detailed Annotation').count()['Chr'].copy()
foo.sort()
foo
HTML(results.to_html())
results = pd.read_table("/home/gpratt/projects/tiffany_genentech/analysis/ad-hoc/LIB19982_SAM18146099_R1_resequenced.R02.polyATrim.adapterTrim.Aligned.out.sorted.rmDup.0x100.peaks_annoated.txt")
foo = results.groupby('Detailed Annotation').count()['Chr'].copy()
foo.sort()
foo
regions = results[7].dropna()
regions = regions.apply(lambda x: x.split()[0])
regions.groupby(regions).count()
HTML(results.to_html())
from Bio import SeqIO
with open("/home/gpratt/Dropbox/Presentations/alus.fasta", 'w') as out_fasta:
for record in SeqIO.parse("/projects/ps-yeolab/genomes/RepBase18.05.fasta/species_specic/homo_sapiens_repbase_fixed.fasta", 'fasta'):
if "alu" in record.name.lower():
SeqIO.write(record, out_fasta, format="fasta")
record.id
# !head /home/gpratt/Dropbox/Presentations/alus.fasta
bar = """Annotation Number of peaks Total size (bp) Log2 Enrichment
3UTR 78.0 12314737 2.613
Other 1.0 2356357 -1.287
Unknown? 0.0 11950 -10.938
RNA 13.0 73904 7.408
miRNA 0.0 16592 -10.938
ncRNA 8.0 2404740 1.684
TTS 37.0 14367940 1.314
LINE 126.0 414468714 -1.768
LINE? 0.0 5789 -10.938
srpRNA 0.0 143444 -10.938
SINE 80.0 216454842 -1.486
RC 0.0 276298 -10.938
tRNA 34.0 65194 8.976
DNA? 0.0 173555 -10.938
pseudo 3.0 831434 1.801
DNA 30.0 61079139 -1.076
Exon 83.0 18265833 2.134
Intron 511.0 380268767 0.376
Intergenic 99.0 545935740 -2.514
Promoter 56.0 15869037 1.769
5UTR 15.0 1389811 3.382
snoRNA 0.0 36 -10.938
LTR? 0.0 12957 -10.938
scRNA 578.0 67815 13.007
CpG-Island 28.0 4878036 2.471
Low_complexity 4.0 9828158 -1.347
LTR 80.0 169888789 -1.137
Simple_repeat 12.0 15139177 -0.386
snRNA 22.0 200066 6.731
Unknown 0.0 838100 -10.938
SINE? 0.0 31483 -10.938
Satellite 0.0 6971418 -10.938
rRNA 64.0 98025 9.300""".split("\n")
#R08
foo = """Annotation Number of peaks Total size (bp) Log2 Enrichment
3UTR 196.0 12314737 3.543
Other 3.0 2356357 -0.101
Unknown? 0.0 11950 -11.337
RNA 20.0 73904 7.631
miRNA 0.0 16592 -11.337
ncRNA 6.0 2404740 0.870
TTS 49.0 14367940 1.321
LINE 230.0 414468714 -1.299
LINE? 0.0 5789 -11.337
srpRNA 2.0 143444 3.352
SINE 343.0 216454842 0.215
RC 0.0 276298 -11.337
tRNA 48.0 65194 9.075
DNA? 0.0 173555 -11.337
pseudo 3.0 831434 1.402
DNA 43.0 61079139 -0.956
Exon 141.0 18265833 2.499
Intron 555.0 380268767 0.096
Intergenic 89.0 545935740 -3.066
Promoter 90.0 15869037 2.054
5UTR 18.0 1389811 3.246
snoRNA 0.0 36 -11.337
LTR? 0.0 12957 -11.337
scRNA 425.0 67815 12.164
CpG-Island 40.0 4878036 2.586
Low_complexity 15.0 9828158 0.161
LTR 44.0 169888789 -2.398
Simple_repeat 79.0 15139177 1.934
snRNA 38.0 200066 7.120
Unknown 0.0 838100 -11.337
SINE? 0.0 31483 -11.337
Satellite 0.0 6971418 -11.337
rRNA 110.0 98025 9.683""".split("\n")
pd.DataFrame([item.split("\t") for item in foo])
pd.DataFrame([item.split("\t") for item in bar])
| 3,443 |
/ComputationNotebooks2015/Wk1_Tue/Probability distributions and random number generation.ipynb | 4fab30c7eaf72357ac8c53727c236936997190a9 | [] | no_license | janice-mccarthy/SummerCourse2016-dev | https://github.com/janice-mccarthy/SummerCourse2016-dev | 1 | 1 | null | null | null | null | Jupyter Notebook | false | false | .r | 971,220 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .r
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: R
# language: R
# name: ir
# ---
# Probability distributions
# ----
#
# To some extent, the foundation of statistics is an understanding of probability distributions. In addition, drawing of random samples from specific probability distributions is ubiquitous in applied statistics and useful in many contexts, not least of which is an appreciation of how different probabilty distributions behave.
help(Distributions)
# Basic functions for working with probabilty distributions
# ----
# In the help documentation, it is stated that "The functions for the density/mass function, cumulative distribution function, quantile function and random variate generation are named in the form dxxx, pxxx, qxxx and rxxx respectively". We will explore what this means with a couple of examples.
# ### Discrete distributions
#
# For discrete distributions, the random values can only take integer values. One of the simplest discrete distributions is the Bernoulli distribution, where the only possible values are 0 ("Failure") and 1 ("Success"). This has 2 parameters - the number of trials $n$ and the probabilty of success in each trial $p$. For example the Bernoulli distribution could model getting HEADs in a sequence of coin tosses, or whether or not a given subject in an experiment responds to a drug. Another discrete distribution is the Binomial distribution, which gives the probability of $k$ successes in $n$ trials. For example, the binomial distibution can tell you how many HEADS you get in 10 coin tosses for a biased coin with p=0.45. Note that the Bernoulli distribution can be considered a special case of the binomial distribution with a size of 1.
?rbinom
# ### The `rxxx` family
# If I coss a toin with probablity of heads = 0.45, how many heads do I see? Let's simulate this experiemnt by drawing random numbers from the Bernoulli distribution (or equivalently the binomial distribution with size=1).
rbinom(n=100, prob=0.45, size=1)
# How many heads do we see if we repeat this experiemnt 10 times?
rbinom(n=10, prob=0.45, size=100)
# It should be clear that the number of heads we observe is alwayss a number between 0 and 100. What is the probabily of observing exactly 35 heades?
# ### The `dxxx` family
dbinom(x=35, prob=0.45, size=100)
# Let's plot the proabbity for all numbers from 0 to 100.
plot(dbinom(x=0:100, prob=0.45, size=100))
# ### The `pxxx` family
# We can also see the cumulative probabilty of otbaiining a certain number of heads with `pbinom`.
plot(pbinom(0:100, prob=0.45, size=100))
# ### The `qxxx` family
# The `qxxx` or quantile function is a little more tricky to understand. One way to think about it is to look at the cumulative distirbution plot and ask - If I start with a horizontal line from the y-axis at some value p between 0 and 1 until I hit the funciton, then drop a veritcal line to teh x-axis, what is the value I get? For example, what is the number of heads where the cumulative distribution first reaches a value of 0.5?
qbinom(p = 0.5, prob = 0.45, size=100)
# Work!
# ----
# What is the mean, median and standard deviation of the number of heads if the probabilty of heads is 0.4 and we toss 50 coins per experiment? Do 1000 such experiments and use the appropirate R functions to callculate these summary statistics.
# +
# -
# What is the probability of getting between 2 to 5 (that is 2, 3, 4 or 5) sixes in 10 rolls of a fair six-sided die?
# +
# -
# Explore what the Poisson distribution is (e.g. see Wikipedia and R help). Suppose there is on averate one mutation every 10,000 bases. What is the probabilty of finding exactly 8 mutations in 100,000 bases? What is the prrobability fo finding 15 or more mutations in 100,000 bases?
# +
# -
# Continuous Distributions
# ----
#
# Distributions can also be continuous - a familiar example is the normal distribution.
x <- seq(-5, 5, length.out = 100)
ns <- rnorm(n=1000, mean=0.0, sd=1.0)
hist(ns, probability=TRUE, ylim = c(0, 1))
lines(x, dnorm(x), type="l", col="blue")
lines(x, pnorm(x), type="l", lty=2, col="red")
# Work!
# ----
# If IQ is normally distributed, the mean IQ is 100 with a standard deviation of 15, and you have an IQ of 135, what percentage of people have IQs higher than you?
# +
# -
# Suppose that you toss 100 unbiased coins per experiment and count the number of heads. Plot the density function of this discrete distribution. Now superimpose a normal distribution density function with mean 50 and standard deviation of 5. What do you observe?
# +
| 4,762 |
/.ipynb_checkpoints/python_sword_for_offer-checkpoint.ipynb | 9ffb8866baee33aa8efd5c4839a154fe0f493fa7 | [] | no_license | Vito-Tu/sword-for-offer | https://github.com/Vito-Tu/sword-for-offer | 7 | 1 | null | null | null | null | Jupyter Notebook | false | false | .py | 315,121 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# <center> ***前言*** </center>
# _以下代码块结合了 [剑指offer][1] 和 [牛客网][2] 来编写,部分代码将在牛客网上进行测试,为了还原剑指offer的算法思想,代码完全仿照C++版进行编写,因此针对python语言还有一定的改进空间_
# [1]https://pan.baidu.com/s/1pavnkxcIRgwR1sKmjosVAQ
# 提取码:za5n
# [2]https://www.nowcode.com/ta/coding-interviews
# + code_folding=[]
# 示例 快速排序法
import random
class Solution:
def Partition(self, data, start, end):
"""随机选择基准数,将较小数移到前方,返回基准数在调整顺序后的索引位置"""
if not data:
raise Exception("Invalid Parameters")
index = random.randrange(start, end)
temp = data[index]
data[index] = data[end]
data[end] = temp
small = start - 1
for index in range(start, end):
if data[index] < data[end]:
small += 1
if small != index:
temp = data[index]
data[index] = data[small]
data[small] = temp
small += 1
temp = data[small]
data[small] = data[end]
data[end] = temp
return small
def QuickSort(self, data, start, end):
"""快速排序法,输入需要排序的范围"""
if start == end:
return
index = self.Partition(data, start, end)
if index > start:
self.QuickSort(data, start, index - 1)
if index < end:
self.QuickSort(data, index + 1, end)
# test code
a = Solution()
data = [1,3,5,76,3,2,6,4]
a.QuickSort(data, 0, len(data)- 1)
data
# -
# # 面试题3:数组中重复的数字
# + 题目一:找出数组中重复的数字
# 在一个长度为n的数组中素有的数字都在[0, n-1]范围内。请找出数组中任意一个重复的数字。例如,输入长度为7的数组{2,3,1,0,2,5,3},那么对应输出的重复数字是2或3.
def duplicate(nums:list):
"""利用列表下标最为哈希表,通过元素互换逐步构建哈希表,若遇到相等的数字则通过下标索引key必然能检测到重复数字
Arg(Argument参数): nums:list[int]
Re(return): 若未找到重复数字,或者列表元素不符合要求将返回bool False,否则返回检索到的重复数字
算法分析:尽管有两重循环,但每个数字最多只要交换两次就能找到属于它的位置,所以总体算法复杂度为O(n)空间复杂度为O(1)
"""
length = len(nums)
if length < 2:
return False
for i in range(length):
if nums[i] < 0 or nums[i] > length - 1:
return False
while nums[i] != i:
if nums[i] != nums[nums[i]]:#如果不相等就互换位置
temp = nums[nums[i]]
nums[nums[i]] = nums[i]
nums[i] = temp
# nums[i], nums[nums[i]] = nums[nums[i] - 1], nums[i] 逗号赋值并非同时进行,在这种情况下无法得到正确结果
else:
return nums[i]
return False
#测试用例
nums = [2,3,1,0,2,5,3]
print(duplicate(nums))
# 牛客网版代码
class Solution:
def duplicate(self, nums, duplication):
"""利用列表下标最为哈希表,通过元素互换逐步构建哈希表,若遇到相等的数字则通过下标索引key必然能检测到重复数字
Arg(Argument参数): nums:list[int]
Re(return): 若未找到重复数字,或者列表元素不符合要求将返回bool False,否则返回检索到的重复数字
算法分析:尽管有两重循环,但每个数字最多只要交换两次就能找到属于它的位置,所以总体算法复杂度为O(n)空间复杂度为O(1)
"""
length = len(nums)
if length < 2:
return False
for i in range(length):
if nums[i] < 0 or nums[i] > length - 1:
return False
while nums[i] != i:
if nums[i] != nums[nums[i]]: # 如果不相等就互换位置
temp = nums[nums[i]]
nums[nums[i]] = nums[i]
nums[i] = temp
else:
duplication[0] = nums[i]
return True
return False
# test code
# 代码已在牛客网通过测试
# + 题目二:(接题目一中)增加限定条件:不修改数组找出重复的数字
# 解题思路:利用二分查找法原理,确定中值,统计数组中比中值小或者比中值大的数字的个数,以此确定重复的数字在中值左侧还是右侧
# +
def countRange(nums, start, end):
"""统计nums中所有在区间[start, end]中的数的个数
"""
if not nums:
return 0
count = 0
for i in range(len(nums)):
if nums[i] >= start and nums[i] <= end:
count += 1
return count
def getDuplication(nums):
"""利用二分查找法来找出数组中任意一重复数字 (注意,此方法不能很好的解决此类问题,偶尔会出现错误)"""
length = len(nums)
if length < 2:
return False
start = 0
end = length - 1
while end >= start:
middle = int((end + start) / 2)
count = countRange(nums, start, middle)
if end == start:
if count > 1:
return start
else:
break
if count > (middle - start + 1):
end = middle
else:
start = middle + 1
return False
#测试用例
nums = [2,4,5,4,3,2,6,7]
print(getDuplication(nums))
# -
# # 面试题 4:二维数组中的查找
# 在一个二维数组中,每一行都按照从左到右的递增顺序排列,每一列都按照从上到下递增的顺序排列,请完成一个函数,输入这样的二维数组和一个整数,判断数组中是否含有该整数.
# 解题思路:首先选取数组右上角的数字,如果该数字大于目标数字则排除该列,小于则排除该行,直到等于或者范围为空停止
def Find(nums, target):
if len(nums) <= 0 or len(nums[0]) <= 0:
return False
i = 0
j = len(nums[0]) - 1
while i <= len(nums) - 1 and j >= 0:
if nums[i][j] > target:
j -= 1
elif nums[i][j] < target:
i += 1
else:
return True
return False
# # 面试题5:替换空格
# 题目:请实现一个函数,把字符串中的每个空格替换成"%20"。例如,输入"We are happy.",则输出"We%20are%20happy."。
# 解题思路:先遍历一遍字符串统计空格的个数,然后分配足够的存储空间,从后向前的依次替换空格(算法时间效率O(n))
# +
def ReplaceBlank(string:list):
"""使用字符型列表来替换str类型,更好的诠释算法思想。若用str类型来做:string.replace(' ', '%20')即可
输入:string:list[char]
输出:string:str
"""
if not string:
return ''
numOfBlank = 0
for i in string:
if i == ' ':
numOfBlank += 1
originalLength = len(string)
newlength = originalLength + 2*numOfBlank
string = string + ['']*2*numOfBlank
while originalLength >= 0 and newlength > originalLength:
if string[originalLength - 1] == ' ':
string[newlength - 1] = '0'
string[newlength - 2] = '2'
string[newlength - 3] = '%'
newlength -= 3
else:
string[newlength - 1] = string[originalLength - 1]
newlength -= 1
originalLength -= 1
return ''.join(string)
string = ['W', 'e', ' ', 'a', 'r', 'e', ' ', 'h', 'a', 'p', 'p', 'y', '.']
print(ReplaceBlank(string))
# -
# # 面试题6: 从尾到头打印链表
# 题目:输入一个链表的头节点,从尾到头反过来打印出每个节点的值。
# 解题思路:利用栈的先进后出特性存储链表然后再输出(或者利用递归函数)
# + code_folding=[7, 19, 36]
#创建链表类
class ListNode:
"""创建链表类"""
def __init__(self, x):
self.val = x
self.next = None
#反转打印函数
def PrintListReversingly_Iteratively(phead):
"""利用栈来实现反向打印链表"""
pNode = phead
stack = []
if pNode.val == None:
print("None!")
while pNode != None:
stack.append(pNode.val)
pNode = pNode.next
while stack != []:
print(stack.pop(-1))
def PrintListReversingly_Recursively(phead):
"""使用递归函数进行反向打印"""
if phead is not None:
if phead.next is not None:
PrintListReversingly_Recursively(phead.next)
print(phead.val)
#测试用例代码
##创建链表
head = ListNode('head->')
pnode = head
valueList = list('abcdefg')
for i in valueList:
pnode.next = ListNode(i)
pnode = pnode.next
#验证链表用代码
'''
pnode = head
while pnode != None:
print(pnode.val)
pnode = pnode.next
print(head.val)
'''
#测试打印函数
PrintListReversingly_Iteratively(head)
PrintListReversingly_Recursively(head)
# + code_folding=[0, 1, 13]
#牛客网解题对应函数(不要在此cell中运行)
def printListFromTailToHead(self, listNode):
'''利用栈实现反向输出链表'''
pNode = listNode
stack = []
if pNode is None:
return stack
while pNode is not None:
stack.append(pNode.val)
pNode = pNode.next
stack.reverse()
return stack
def printListFromTailToHead(self, listNode):
'''利用递归函数反向输出链表'''
if listNode is None:
return []
else:
return self.printListFromTailToHead(listNode.next) + [listNode.val]
# + [markdown] heading_collapsed=true
# # 面试题7:重建二叉树
# 题目:输入某二叉树的前序遍历和中序遍历的结果,请重建该二叉树。假设输入的前序遍历和中序遍历的结果中都不含重复数字。例如输入前序遍历序列{1,2,4,7,3,5,6,8}和中序遍历序列{4,7,2,1,5,3,8,6},则重建如下入的二叉树并输出它的头节点。
# 
# + hidden=true
class BinaryTreeNode:
'''创建二叉树节点'''
def __init__(self, x=None):
self.val = x
self.left = None
self.right = None
def preTree(treehead):
"""递归的先根序遍历二叉树,返回列表序列"""
if treehead is None:
return []
return [treehead.val] + preTree(treehead.left) + preTree(treehead.right)
def tinTree(treehead):
"""递归的中根序遍历二叉树,返回列表序列"""
if treehead is None:
return []
return tinTree(treehead.left) + [treehead.val] + tinTree(treehead.right)
def _ConstructBinaryTree(pre,tin):
"""递归调用构造子二叉树"""
root = BinaryTreeNode(pre[0])
#递归终止条件
if len(pre) == 1:
if len(tin) == 1 and pre[0] == tin[0]:
return root
else:
raise Exception("input error")
rootInorderIndex = tin.index(pre[0])
if len(tin) == 1 and tin[rootInorderIndex] != root.val:
raise Exception("input error")
left = tin[0:rootInorderIndex]
right = tin[rootInorderIndex+1:]
if len(left) > 0:
root.left = _ConstructBinaryTree(pre[1:len(left) + 1], left)
if len(right) > 0:
root.right = _ConstructBinaryTree(pre[len(left)+1:], right)
return root
def reConstructBinaryTree(pre, tin):
"""初始判断输入是否合法,并递归调用构造函数"""
if not pre or not tin:
raise Exception("input error")
return _ConstructBinaryTree(pre, tin)
#测试代码
prelist = [1,2,4,7,3,5,6,8]
tinlist = [4,7,2,1,5,3,8,6]
treehead = reConstructBinaryTree(prelist, tinlist)
#验证prelist和tinlist
print(tinTree(treehead))
# + hidden=true
#牛客网通过代码 (#####!!不要运行此段代码!!#####)
def _ConstructBinaryTree(self,pre,tin):
root = TreeNode(pre[0])
if len(pre) == 1:
if len(tin) == 1 and pre[0] == tin[0]:
return root
else:
return False
rootInorderIndex = tin.index(pre[0])
if len(tin) == 1 and tin[rootInorderIndex] != root.val:
return False
left = tin[0:rootInorderIndex]
right = tin[rootInorderIndex+1:]
if len(left) > 0:
root.left = self._ConstructBinaryTree(pre[1:len(left) + 1], left)
if len(right) > 0:
root.right = self._ConstructBinaryTree(pre[len(left)+1:],right)
return root
def reConstructBinaryTree(self, pre, tin):
if not pre or not tin:
return None
return self._ConstructBinaryTree(pre, tin)
# + [markdown] heading_collapsed=true
# # 面试题8:二叉树的下一个节点
# 题目:给定一颗二叉树和其中的一个节点,如何找出中序遍历序列的下一个节点?树中的节点除了有两个分别指向左、右子节点的指针,还有一个指向父节点的指针。
# + code_folding=[] hidden=true
class BinaryTreeNode:
'''创建二叉树节点'''
def __init__(self, x=None):
self.val = x
self.parent = None
self.left = None
self.right = None
def GetNext(pNode):
"""获取"""
if pNode == None:
return None
pNext = BinaryTreeNode()
if pNode.right != None:
#若存在右子树,则右子树中最左侧的节点是下一节点
pRight = pNode.right
while pRight.left != None:
pRight = pRight.left
pNext = pRight
elif pNode.parent != None:
#若右子树不存在,则父节点中是左子树的父节点是下一节点
pCurrent = pNode
pParent = pNode.parent
while pParent != None and pCurrent == pParent.right:
pCurrent = pParent
pParent = pParent.parent
pNext = pParent
return pNext
#test code
#tin=list('dbheiafcg')
#pre=list('abdehicfg')
headNode = BinaryTreeNode('a')
headNode.left = BinaryTreeNode('b')
headNode.left.parent = headNode
pNode = headNode.left
pNode.left = BinaryTreeNode('d')
pNode.right = BinaryTreeNode('e')
pNode.left.parent = pNode
pNode.right.parent = pNode
pNode = pNode.right
pNode.left = BinaryTreeNode('h')
pNode.right = BinaryTreeNode('i')
pNode.left.parent = pNode
pNode.right.parent = pNode
headNode.right = BinaryTreeNode('c')
headNode.right.parent = headNode
pNode = headNode.right
pNode.left = BinaryTreeNode('f')
pNode.right = BinaryTreeNode('g')
pNode.left.parent = pNode
pNode.right.parent = pNode
result = GetNext(pNode)
print(result.val)
# + hidden=true
# 牛客网版代码
class Solution:
def GetNext(self, pNode):
if pNode == None:
return None
pNext = None
if pNode.right != None:
# 若存在右子树,则右子树中最左侧的节点是下一节点
pRight = pNode.right
while pRight.left != None:
pRight = pRight.left
pNext = pRight
elif pNode.next != None:
# 若右子树不存在,则父节点中是左子树的父节点是下一节点
pCurrent = pNode
pParent = pNode.next
while pParent != None and pCurrent == pParent.right:
pCurrent = pParent
pParent = pParent.next
pNext = pParent
return pNext
# test code
# 代码已在牛客网通过测试
# -
# # 面试题9:用两个栈实现队列
# 题目:用两个栈实现一个队列。队列的声明如下,请实现它的两个函数appendTail和deleteHead,分别完成在队列尾部插入节点在队列头部删除节点的功能。
# 解题思路:利用栈1入队,在栈2为空时将栈1中的元素通过出栈,再入栈的方式转移到栈2中实现反序
class Solution:
def __init__(self):
self.stack1 = []
self.stack2 = []
def push(self, node):
self.stack1.append(node)
def pop(self):
if len(self.stack2) <= 0:
#若栈2为空则转移已入栈元素
while len(self.stack1) > 0:
self.stack2.append(self.stack1.pop())
if len(self.stack2) == 0:
#若已转移元素之后栈2还是空,则说明队列为空
raise Exception('queue is empty')
return self.stack2.pop()
#测试代码
inqueue = [1,2,3,4,5,6,7,8,9]
testqueue = Solution()
for i in inqueue:
testqueue.push(i)
while testqueue.stack1 or testqueue.stack2:
print(testqueue.pop())
# # 面试题10:斐波拉契数列
# 题目1:求斐波拉契数列的第n项。
# [斐波拉契数列定义] f(n) = f(n-1) + f(n-2)、f(0) = 0、f(1) = 1
#
# 解题思路:可以用递归的方式来实现,但算法效率低,使用循环来代替递归会更好
#
# 题目二:青蛙跳台阶问题
# 一只青蛙一次可以跳上一级台阶,也可以跳上2级台阶。求该青蛙跳上一个n级台阶总共有多少种跳法。
#
# 解题思路:f(1) = 1, f(2) = 2, 一般情况,先跳一级,剩余还有f(n-1)种方法,先跳2级,剩余还有f(n-2)种方法,所以f(n) = f(n-1) + f(n-2),因此与斐波拉契数列相似
# + code_folding=[0, 15, 22, 37]
def Fibonacci(n):
"""从小到大计算,类推出第n项,时间复杂度O(n)比递归小"""
result = [0,1]
if n < 2:
return result[n]
fibOne = 1
fibTwo = 0
temp = None
for i in range(2,n+1):
temp = fibOne + fibTwo
fibTwo = fibOne
fibOne = temp
return temp
Fibonacci(10)
def FibonacciM():
"""利用数学公式[[f(n), f(n-1)], [f(n-1), f(n-2)]] = [[1, 1], [1,0]]^(n-1)。此方法时间复杂度为O(log(n))"""
print("此函数待实现,留作练习")
pass
"""函数Fibonacci在牛客网上测试通过"""
class Solution:
"""青蛙台阶问题、矩形覆盖问题"""
def jumpFloor(self, number):
result = [0,1,2]
if number < 3:
return result[number]
fibOne = 2
fibTwo = 1
fibN = None
for i in range(3,number+1):
fibN = fibOne + fibTwo
fibTwo = fibOne
fibOne = fibN
return fibN
class Solution:
"""青蛙跳台阶变态版:青蛙每次可以跳任意级"""
def jumpFloorII(self, number):
if number == 0:
return 0
return 2**(number - 1)
# + [markdown] heading_collapsed=true
# # 面试题11:旋转数组中的最小数字
# 题目:把一个数组最开始的若干个元素搬到数组的末尾,我们称之为数组的旋转。输入一个递增排序的数组的一个旋转,输出旋转数组的最小元素。例如,数组[3,4,5,1,2]为[1,2,3,4,5]的一个旋转,该数组的最小值为1.
#
# 解题思路:旋转后的排序数组是由两个已排好序的子数组组成,利用对撞指针的二分查找法逐步缩小查找范围,但若遇上大量重复数字时,需要遍历子数组进行查找
#
# + hidden=true
def MinInorder(nums):
"""顺序查找最小数字子函数"""
result = nums[0]
for i in nums:
if i < result:
result = i
return result
def Min(nums):
if not nums:
raise Exception("无效的输入")
length = len(nums)
if length < 2:
return nums[0]
start = 0
end = length - 1
mid = start
while nums[start] >= nums[end]:
if end - start == 1:
mid = end
break
mid = (end + start) / 2
if nums[start] == nums[end] and nums[mid] == nums[start]:
return MinInorder(nums[start:end])
if nums[mid] >= nums[start]:
start = mid
elif nums[mid] <= nums[end]:
end = mid
return nums[mid]
# test code
"""在牛客网上通过测试"""
# -
# # 面试题12:矩阵中的路径
# 题目:请设计一个函数,用来判断在一个矩阵中是否存在一条包含某字符串的所有字符的路径。路径可以从矩阵中的任意一格开始,每一步可以在矩阵中向左、右、上、下移动一格。如果一条路径经过了矩阵的某一格,那么该路径不能再次进入该格子。例如,在下面的3*4的矩阵中包含一条字符串“bfce”的路径(路径中的字母用大写标出)。但矩阵中不包含字符串“abfb”的路径,因为字符串的第一个字符b占据了矩阵中的第一行第二个格子后,路径不能再次进入这个格子。
# [[a,B,t,g]
# ,[c,F,C,s]
# ,[j,d,E,h]]
#
# 解题思路:利用回溯法,若当前路径不通,则回溯上一节点,尝试未尝试过的路径
# + code_folding=[]
def hasPathCore(matrix, i, j, strs, visited, pathLength):
"""递归调用自己,探索每一个节点"""
if pathLength >= len(strs):
return True
result = False
if i >= 0 and i < len(matrix) and j >= 0 and j < len(matrix[0]) and matrix[i][j] == strs[pathLength] and not visited[i][j]:
# 索引范围正确且此时矩阵的值和字符串的值相匹配,并且此点未被占据过,则递归探索下一符合要求的点
pathLength += 1
visited[i][j] = True
result = hasPathCore(matrix,i,j-1,strs,visited,pathLength) or hasPathCore(matrix,i-1,j,strs,visited,pathLength) \
or hasPathCore(matrix,i,j+1,strs,visited,pathLength) or hasPathCore(matrix,i+1,j,strs,visited,pathLength)
# 递归探索四个方向的下一节点是否符合要求,若不符合要求则退回上一节点
if not result:
pathLength -= 1
visited[i][j] = False
return result
def hasPath(matrix,strs):
if not matrix or not strs:
return False
rows = len(matrix)
clos = len(matrix[0])
# visited = [[False]*clos]*rows
# 这样构造矩阵,其行将会是第一行的浅拷贝,修改任一元素,将引起整列元素同时被改,因此采用下面的循环进行构造
visited = [[False]*clos for _ in range(rows)]
pathLength = 0
for i in range(rows):
for j in range(clos):
#双循环遍历每一个点作为起点
if hasPathCore(matrix,i,j,strs,visited,pathLength):
return True
return False
#test code
a = [[None]]*3
a[0] = list('abtg')
a[1] = list('cfcs')
a[2] = list('jdeh')
strs = 'tegsc'
print(hasPath(a, strs))
# -
# 牛客网版代码
class Solution:
def hasPathCore(self, matrix, i, j, strs, visited, pathLength):
"""递归调用自己,探索每一个节点"""
if pathLength >= len(strs):
return True
result = False
if i >= 0 and i < len(matrix) and j >= 0 and j < len(matrix[0]) and matrix[i][j] == strs[pathLength] and not visited[i][j]:
# 索引范围正确且此时矩阵的值和字符串的值相匹配,并且此点未被占据过,则递归探索下一符合要求的点
pathLength += 1
visited[i][j] = True
result = self.hasPathCore(matrix,i,j-1,strs,visited,pathLength) or self.hasPathCore(matrix,i-1,j,strs,visited,pathLength) or self.hasPathCore(matrix,i,j+1,strs,visited,pathLength) or self.hasPathCore(matrix,i+1,j,strs,visited,pathLength)
# 递归探索四个方向的下一节点是否符合要求,若不符合要求则退回上一节点
if not result:
pathLength -= 1
visited[i][j] = False
return result
def hasPath(self, matrix, rows, cols, strs):
if not matrix or not strs:
return False
temp = [list(matrix[i:i+cols]) for i in range(0, len(matrix), cols)]
# visited = [[False]*clos]*rows
# 这样构造矩阵,其行将会是第一行的浅拷贝,修改任一元素,将引起整列元素同时被改,因此采用下面的循环进行构造
visited = [[False]*cols for _ in range(rows)]
pathLength = 0
for i in range(rows):
for j in range(cols):
#双循环遍历每一个点作为起点
if self.hasPathCore(temp,i,j,strs,visited,pathLength):
return True
return False
# test code
# 代码已在牛客网通过测试
# + [markdown] heading_collapsed=true
# # 面试题13:机器人的运动范围
# 题目:地上有一个m×n的方格。一个机器人从坐标(0, 0)的格子开始移动,它每次可以向左、右、上、下移动一格,但不能进入行坐标和列坐标的位数之和大于k的格子。例如,当k=18时,机器人能够进入方格(35,37),因为3+5+3+7=18。但它不能进入方格(35, 38),因为3+5+3+8=19。请问该机器人能够到达多少个格子?
#
# 解题思路:因为机器人每次只能移动一格,同样可以利用回溯法逐步遍历能够到达哪些格子
# + code_folding=[0, 9, 17, 29] hidden=true
def getDigitSum(number):
"""计算数字和函数"""
sumResult = 0
while number > 0:
sumResult += number % 10
number /= 10
return sumResult
def check(threshold, rows, cols, row, col, visited):
"""条件判断函数"""
if row >= 0 and row < rows and col >= 0 and col < cols and getDigitSum(row) + getDigitSum(col)\
<= threshold and not visited[row][col]:
return True
return False
def movingCountCroe(threshold, rows, cols, row, col, visited):
"""递归探索每一个可能的格子"""
count = 0
if check(threshold, rows, cols, row, col, visited):
visited[row][col] = True
count = 1 + movingCountCroe(threshold, rows, cols, row - 1, col, visited) \
+ movingCountCroe(threshold, rows, cols, row, col - 1, visited)\
+ movingCountCroe(threshold, rows, cols, row + 1, col, visited)\
+ movingCountCroe(threshold, rows, cols, row, col + 1, visited)
return count
def movingCount(threshold, rows, cols):
"""主函数,初始化visited方便核心函数递归调用"""
if threshold <= 0 or rows <= 0 or cols <= 0:
return 0
visited = [[False]*cols for _ in range(rows)]
count = movingCountCroe(threshold, rows, cols, 0, 0, visited)
return count
# test code
print(movingCount(18, 35, 35))
# + hidden=true
# 牛客网版代码
class Solution:
def getDigitSum(self, number):
"""计算数字和函数"""
sumResult = 0
while number > 0:
sumResult += number % 10
number /= 10
return sumResult
def check(self, threshold, rows, cols, row, col, visited):
"""条件判断函数"""
if row >= 0 and row < rows and col >= 0 and col < cols and self.getDigitSum(row) + self.getDigitSum(col) \
<= threshold and not visited[row][col]:
return True
return False
def movingCountCroe(self, threshold, rows, cols, row, col, visited):
"""递归探索每一个可能的格子"""
count = 0
if self.check(threshold, rows, cols, row, col, visited):
visited[row][col] = True
count = 1 + self.movingCountCroe(threshold, rows, cols, row - 1, col, visited) \
+ self.movingCountCroe(threshold, rows, cols, row, col - 1, visited) \
+ self.movingCountCroe(threshold, rows, cols, row + 1, col, visited) \
+ self.movingCountCroe(threshold, rows, cols, row, col + 1, visited)
return count
def movingCount(self, threshold, rows, cols):
"""主函数,初始化visited方便核心函数递归调用"""
if threshold <= 0 or rows <= 0 or cols <= 0:
return 0
visited = [[False] * cols for _ in range(rows)]
count = self.movingCountCroe(threshold, rows, cols, 0, 0, visited)
return count
# test code
# 代码已在牛客网通过测试
# + [markdown] heading_collapsed=true
# # 面试题14:剪绳子
# 题目:给你一根长度为n的绳子,请把绳子剪成m段(m,n都是整数,n>1并且m>1),每段绳子的长度记为k[0],k[1],...,k[m]。请问k[0]*k[1]*...*k[m]可能的最大乘积是多少?例如,当绳子的长度为8时,我们把他剪成长度分别为2,3,3的三段,此时得到的最大乘积是18.
#
# 解题思路:因为是最优化问题,且问题可分,可考虑使用动态规划或者贪婪法,详细见代码文档或剑指offer(注:题目中需补充条件,m为任意整数,且切成的绳子长度必须为整数)
# + code_folding=[0, 24] hidden=true
def maxProductAfterCutting_solution(length):
"""动态规划法,从上而下的分析分解问题,自下而上的顺序计算问题"""
result = [0, 1, 2, 3]
if length < 2:
return 0
elif length < 4:
return result[length - 1]
products = [None]*(length + 1)
for i in result:
products[i] = i
maxValue = 0
for i in range(4, length + 1):
maxValue = 0
j = 1
while j <= i/2:
product = products[j]*products[i-j]
if maxValue < product:
maxValue = product
products[i] = maxValue
j += 1
maxValue = products[length]
return maxValue
def maxProductAfterCutting_solution2(length):
"""贪婪算法,尽可能多的剪长度为3的绳子,当剩下的绳子长度为4时,把绳子剪成两段为2的绳子。
解释:当n>=5时,3*(n-3)>=2*(n-2)>n,故选用上述策略"""
result = [0, 1, 2, 3]
if length < 2:
return 0
elif length < 4:
return result[length - 1]
timesOf3 = length / 3
if length - timesOf3 * 3 == 1:
timesOf3 -= 1
timesOf2 = (length - timesOf3 * 3) / 2
return int(pow(3, timesOf3))*int(pow(2, timesOf2))
# test code
print(maxProductAfterCutting_solution(8))
print(maxProductAfterCutting_solution2(8))
# + [markdown] heading_collapsed=true
# # 面试题15:二进制中1的个数
# 题目:请实现一个函数,输入一个整数,输出该数的二进制表示中1的个数。例如,把9表示成二进制是1001,有2位是1,因此,输入9,则函数输出2.
# + hidden=true
def NumberOf1Croe(n):
"""将1逐位与n按位与,检测该二进制位是否有1,若有则计数。该算法需要循环sizeof(int)次"""
count =0
flag = 1
while flag <= abs(n):
if n&flag:
count += 1
flag = flag << 1
return count
def NumberOf1(n):
"""计算整数的二进制数中1的个数,由于在python中负数没有用补码表示,故在主函数中另行处理"""
if n < 0:
return 32 - NumberOf1Croe(abs(n) -1)
else:
return NumberOf1Croe(n)
def numberOf1Croe(n):
"""将一个整数减1,都是把二进制中最右边的1变成0,并且把它右边的所有0变成1,将此数再与原数按位与,相当于将原数二进制数中最右
边的1变成0,整数中有多少个1就可以做多少次这种操作。基于这种思想写出类的算法需要循环count次,即有多少个1循环多少次"""
count =0
while n:
count += 1
n = (n-1)&n
return count
def numberOf1(n):
"""计算整数的二进制数中1的个数,由于在python中负数没有用补码表示,故在主函数中另行处理"""
if n <0:
return 32 - numberOf1Croe(abs(n) -1)
else:
return numberOf1Croe(n)
#test code
print(NumberOf1(-3))
print(numberOf1(-3))
"""
#将int类型转为补码表示的函数
def int2Bin32(i):
return (bin(((1<<32) - 1) & i)[2:]).zfill(32)
"""
# + [markdown] heading_collapsed=true
# # 面试题16:数值的整数次方
# 题目:实现函数double Power(double base, int exponent),求base的exponent次方。不得使用库函数,同时不需要考虑大数问题。
# + hidden=true
def PowerWithUnsignedExponent(base, exponent):
"""使用循环连乘来计算无符号指数"""
result = 1.0
for i in range(exponent):
result *= base
return result
def PowerWithUnsignedExponent2(base, exponent):
"""递归的将指数拆分成平方,减少乘法的次数,并用右移运算符代替除2,利用按位与运算符来判断函数奇偶"""
if exponent ==0:
return 1
if exponent == 1:
return base
result = PowerWithUnsignedExponent2(base, exponent >> 1)
result *= result #平方
if exponent & 0x1 == 1:
result *=base
return result
g_InvalidInput = False
#错误输入指示,在牛客网python编程中,请删除此功能,否则不能通过测试
def Power(base, exponent):
"""主函数。浮点型数据类型不可直接用‘==’判断是否为0,可借助比较较小数1.0e-9来判断"""
g_InvalidInput = False
if abs(base) < 1.0e-9 and exponent < 0:
g_InvalidInput = True
return 0.0
absExponent = abs(exponent)
result = PowerWithUnsignedExponent2(base, absExponent)
if exponent < 0:
result = 1.0/result
return result
#test code
base = 2
exponent = 10
print(Power(base, exponent))
# + [markdown] heading_collapsed=true
# # 面试题17:打印从1到最大的n位数
# 题目:输入数字n,按顺序打印出从1到最大的n位十进制数。比如输入3,则打印输出1,2,3一直到最大的3为数999.
#
# 解题思路:此题涉及到大数问题,python内置类型智能支持大数,但为了表现算法思想不使用python内置大数,转而使用list[str]来表示大数
# + hidden=true
def PrintNumber(nums):
"""打印函数,从第一个非零数字开始打印,循环查找第一个非零数字打印并跳出循环
nums必须是list类型"""
isBeginning = True
for i in range(len(nums)):
if isBeginning and nums[i] != '0':
isBeginning = False
if not isBeginning:
print(''.join(nums[i:]))
break
def Increment(nums):
"""数字自增函数,并判断是否溢出,返回溢出标志,nums必须是list[str]类型,list是可变类型,在传参的时候类似于c++
中传指针,所以无需返回即可修改传入nums中的元素"""
isOverflow = False
nTakeOver = 0
nLength = len(nums)
for i in range(nLength - 1,-1,-1):
#反向循环[nLength,0],设置步长为-1即反向
nSum = ord(nums[i]) - ord('0') + nTakeOver
if i == nLength - 1:
nSum += 1
if nSum >= 10:
if i == 0:
isOverflow = True
else:
nSum -= 10
nTakeOver = 1
nums[i] = chr(ord('0') + nSum)
else:
nums[i] = chr(ord('0') + nSum)
break
return isOverflow
def PrintToMaxOfNDigits(n):
"""打印主函数,生成长度为n的list[str]来表示n位数字,可表示任意大的数字"""
if n <= 0:
return
nums = ['0']*n
while not Increment(nums):
PrintNumber(nums)
def PrintToMaxOfNDigitsRecursively(nums, length, index):
"""利用排列组合的思想,递归的循环调用函数,打印不同的数字组合"""
if index == length - 1:
PrintNumber(nums)
return
for i in range(10):
nums[index+1] = chr(ord('0') + i)
PrintToMaxOfNDigitsRecursively(nums, length, index + 1)
def PrintToMaxOfNDigits2(n):
"""打印数字,递归调用版"""
if n <=0:
return
nums = ['0']*n
for i in range(10):
nums[0] = chr(i+ord('0'))
PrintToMaxOfNDigitsRecursively(nums, len(nums), 0)
#test code
PrintToMaxOfNDigits(-1)
PrintToMaxOfNDigits2(1)
# -
# # 面试题18:删除链表的节点
# + 题目一:在O(1)时间内删除链表节点。
# 给定单向链表的头指针和一个节点指针,定义一个函数在O(1)时间内删除该节点。
# +
class ListNode:
"""创建链表类"""
def __init__(self, x=None):
self.val = x
self.next = None
def DeleteNode(pListHead, pToBeDelete):
"""通过复制的方式删除指定节点,若要删除尾节点,则只能遍历再删除。平均时间复杂度[(n-1)*O(1)+O(n)]/n=O(1)"""
# print("pListHead's id is: "+str(id(pListHead)))
# print("pToBeDelete's id is: "+str(id(pToBeDelete)))
if not pListHead or not pToBeDelete:
return
if pToBeDelete.next != None:
pNext = pToBeDelete.next
pToBeDelete.val = pNext.val
# print("pToBeDelete.val's id is: "+str(id(pToBeDelete.val)))
pToBeDelete.next = pNext.next
elif pListHead == pToBeDelete:
pListHead.val = None
pListHead.next = None
# pListHead = ListNode()
# print("pListHead's id is: "+str(id(pListHead)))
else:
pNode = pListHead
while pNode.next != pToBeDelete:
pNode = pNode.next
pNode.next = None
# return pListHead
##创建链表
head = ListNode('head->')
# print("head's id is: "+str(id(head)))
#可变对象直接赋值就是浅拷贝,相当于c++中传递地址
pnode = head
# print("pnode's id is: "+str(id(pnode)))
valueList = list('')
for i in valueList:
pnode.next = ListNode(i)
pnode = pnode.next
# print("pnode's id is: "+str(id(pnode)))
# gnode = ListNode('1')
DeleteNode(head, pnode)
#验证链表用代码
# print(gnode.val)
rnode = head
while rnode != None:
print(rnode.val)
rnode = rnode.next
"""
!!!注:python传参与c++中常引用传参相似,对于可变对象,不可直接改变对象本身,但可改变对象内置的值,如直接
改链表头不可行,但可以改链表头的值及其他未传入的节点,若一定要改链表头则只能通过返回值的方式进行,例如list对象
"""
# + 题目二:删除链表中重复的节点。
#该算法仅能删除相邻重复节点,比如[1,2,3,4,1]首位节点重复就不会被删除,故不作深入测试
class Solution:
def deleteDuplication(self, phead):
if phead == None:
return phead
pPreNode = None
pNode = phead
while pNode != None:
pNext = pNode.next
needDelete = False
if pNext != None and pNext.val == pNode.val:
needDelete = True
if not needDelete:
pPreNode = pNode
pNode = pNode.next
else:
value = pNode.val
pToBeDel = pNode
while pToBeDel != None and pToBeDel.val == value:
pNext = pToBeDel.next
del pToBeDel
pToBeDel = pNext
if pPreNode == None:
phead = pNext
else:
pPreNode.next = pNext
pNode = pNext
return phead
# test code
# 代码已在牛客网通过测试
# + [markdown] heading_collapsed=true
# # 面试题19:正则表达式匹配
# 题目:请实现一个函数用来匹配包含"."和"\*"的正则表达式。模式中的字符"."表示任意一个字符,而"\*"表示它前面的字符可以出现任意次(含0次)。在本题中,匹配是指字符串的所有字符匹配整个模式。例如,字符串"aaa"与模式"a.a"和"ab\*ac\*a"匹配,但与"aa.a"和"ab\*a"均不匹配。
# + hidden=true
class Solution:
def matchCore(self, strs, pattern):
"""递归探索每一种情况"""
if not strs and not pattern:
return True
if strs and not pattern:
return False
if len(pattern) > 1 and pattern[1] == '*':
if strs and (pattern[0] == strs[0] or pattern[0] == '.'):
return self.matchCore(strs[1:],pattern[2:]) or self.matchCore(strs[1:],pattern) or self.matchCore(strs, pattern[2:])
else:
return self.matchCore(strs, pattern[2:])
if strs and (strs[0] == pattern[0] or pattern[0] == '.'):
return self.matchCore(strs[1:], pattern[1:])
return False
def match(self, s, pattern):
if s == None or pattern == None:
return False
return self.matchCore(s, pattern)
a = Solution()
print(a.match('aaa', 'a.a'))
# + [markdown] heading_collapsed=true
# # 面试题20:表示数值的字符串
# 题目:请实现一个函数用来判断字符串是否表示数值(包括整数和小数)。例如,字符串"+100","5e2","-123","3.1416","-1E-16"都表示数值,但"12e","1a3.14","1.2.3","+-5","12e+5.4"都不是
# + hidden=true
class Solution:
def scanUnsignedInterger(self, strs):
"""对传入字符串从头计算是无符号数字的长度,返回长度"""
if not strs:
return 0
i = 0
while i >= 0 and i < len(strs) and strs[i] >= '0' and strs[i] <= '9':
i += 1
return i
def scanInteger(self, strs):
"""对传入字符串从头计算有符号数字的长度,返回长度"""
if not strs:
return 0
if strs[0] in '-+':
return 1 + self.scanUnsignedInterger(strs[1:])
return self.scanUnsignedInterger(strs)
def isNumeric(self, s):
"""使用index表示首个未进行遍历的字符的索引,使用temp变量表示上一个匹配的长度,为应对index有可能超出字符串长度,
每次进行判断之前先进行判断是否溢出"""
if not s:
return False
index = 0
temp = self.scanInteger(s)
numeric = temp > 0
index += temp
if index < len(s) and s[index] == '.':
index += 1
temp = self.scanUnsignedInterger(s[index:])
numeric = temp or numeric
index += temp
if index < len(s) and (s[index] == 'e' or s[index] == 'E'):
index += 1
temp = self.scanInteger(s[index:])
numeric = numeric and temp
index += temp
return bool(numeric) and index == len(s)
#test code
a_test = Solution()
print(a_test.isNumeric('+100'))
# + [markdown] heading_collapsed=true
# # 面试题21:调整数组顺序使奇数位于偶数前面
# 题目:输入一个整数数组,实现一个函数来调整该数组中数字的顺序,使得所有的奇数位于数组的前半部分,所有的偶数位于数组的后半部分。
# + hidden=true
class Solution:
def reOrderArray(self, array):
"""对撞指针法,发现不符合要求的元素则与另一不符合要求的元素调换顺序"""
if not array:
return
pBegin = 0
pEnd = len(array) - 1
while pBegin < pEnd:
while pBegin < pEnd and array[pBegin] & 0x1 != 0:
pBegin += 1
while pBegin < pEnd and array[pEnd] & 0x1 == 0:
pEnd -= 1
if pBegin < pEnd:
temp = array[pBegin]
array[pBegin] = array[pEnd]
array[pEnd] = temp
#test code
a_test = Solution()
b_list = [1,2,3,4,5,6,7,8,9]
a_test.reOrderArray(b_list)
print(b_list)
# + hidden=true
class Solution:
"""将判断函数解耦分离出来,可以让程序具有更好的通用性"""
def _isEven(self, n):
"""判断函数"""
return (n&1) == 0
def reOrderArray(self, array):
"""主函数,指针对撞,交换不符合条件的值"""
if not array:
return
pBegin = 0
pEnd = len(array) - 1
while pBegin < pEnd:
while pBegin < pEnd and not self._isEven(array[pBegin]):
pBegin += 1
while pBegin < pEnd and self._isEven(array[pEnd]):
pEnd -= 1
if pBegin < pEnd:
temp = array[pBegin]
array[pBegin] = array[pEnd]
array[pEnd] = temp
#test code
a_test = Solution()
b_list = [1,2,3,4,5,6,7,8,9]
a_test.reOrderArray(b_list)
print(b_list)
# + hidden=true
#注意牛客网上增加了附加条件,奇数与奇数之间,偶数与偶数之间的相对位置不能改变,目前暂无较好的解法
class Solution:
def reOrderArray(self, array):
# write code here
count = 0
for i in range(0,len(array)):
for j in range(len(array)-1,i,-1):
if array[j-1]%2 ==0 and array[j]%2==1:
array[j], array[j-1]= array[j-1], array[j]
return array
# + [markdown] heading_collapsed=true
# # 面试题22:链表中倒数第K个节点
# 题目:输入一个链表,输出该链表中倒数第k个节点。为了符合大多数人的习惯,本题从1开始计数,即链表的尾节点是倒数第一个节点。例如,一个链表有6个节点,从头结点开始,它们的值依次是1,2,3,4,5,6。这个链表的倒数第三个节点是值为4的节点。
# + hidden=true
class ListNode:
"""创建链表类"""
def __init__(self, x=None):
self.val = x
self.next = None
class Solution:
def FindKthToTail(self, head, k):
if head is None or k <= 0:
return None
pAHead = head
for i in range(k-1):
if pAHead.next != None:
pAHead = pAHead.next
else:
return None
pBHead = head
while pAHead.next != None:
pAHead = pAHead.next
pBHead = pBHead.next
return pBHead
#test code
#牛客网已通过测试
# + [markdown] heading_collapsed=true
# # 面试题23:链表中环的入口节点
# 题目:如果一个链表中包含环,如何找出环的入口节点?例如在如图3.8所示的链表中,环的入口节点是3
# 1->2->3->4->5->6 and 6->3
# + hidden=true
class Solution:
def MeetingNode(self, pHead):
"""利用速度不同的双指针,检查链表中是否包含环,若包含则返回环中的节点"""
if pHead == None:
return None
pSlow = pHead.next
if pSlow == None:
return None
pFast = pSlow.next
while pFast != None and pSlow != None:
if pFast == pSlow:
return pFast
pSlow = pSlow.next
pFast = pFast.next
if pFast != None:
pFast = pFast.next
return None
def EntryNodeOfLoop(self, pHead):
"""获取环中的节点,并循环统计环中包含节点的个数nodesInLoop,再利用间隔nodesInLoop个节点的指针对撞,判断入口"""
meetingNode = self.MeetingNode(pHead)
if meetingNode == None:
return None
nodesInLoop = 1
pNode1 = meetingNode
while pNode1.next != meetingNode:
pNode1 = pNode1.next
nodesInLoop += 1
pNode1 = pHead
for i in range(nodesInLoop):
pNode1 = pNode1.next
pNode2 = pHead
while pNode1 != pNode2:
pNode1 = pNode1.next
pNode2 = pNode2.next
return pNode1
#test code
#牛客网上已通过测试
# + [markdown] heading_collapsed=true
# # 面试题24:反转链表
# 题目:定义一个函数,输入一个链表的头节点,反转该链表并输出反转后的链表的头结点。
# + hidden=true
class Solution:
# 返回ListNode
def ReverseList(self, pHead):
"""利用三个指针分别指向当前指针的前和后节点,以防止链表反转时断裂,并初始化反转后链表的头节点和前节点,
以防返回出错"""
pReversedHead = None
pNode = pHead
pPrev = None
while pNode != None:
pNext = pNode.next
if pNext == None:
pReversedHead = pNode
pNode.next = pPrev
pPrev = pNode
pNode = pNext
return pReversedHead
#test code
#代码通过牛客网测试
# + [markdown] heading_collapsed=true
# # 面试题25:合并两个排序的链表
# 题目:输入两个递增排序的链表,合并这两个链表并使新链表中的节点仍然是递增排序的。
# + hidden=true
class Solution:
# 返回合并后列表
def Merge(self, pHead1, pHead2):
"""递归比较每一个节点的值,并将较小的节点加入新的链表中"""
if pHead1 == None:
return pHead2
elif pHead2 == None:
return pHead1
pMergedHead = None
if pHead1.val < pHead2.val:
pMergedHead = pHead1
pMergedHead.next = self.Merge(pHead1.next, pHead2)
else:
pMergedHead = pHead2
pMergedHead.next = self.Merge(pHead1, pHead2.next)
return pMergedHead
#test code
#牛客网已通过测试
# + [markdown] heading_collapsed=true
# # 面试题26:树的子结构
# 题目:输入两颗二叉树A和B,判断B是不是A的子结构。
# + hidden=true
class Solution:
def Equal(self, num1, num2):
"""定义通用的比较函数,可以兼容更多不同类型的存储值"""
if abs(num1 - num2) < 1E-7:
return True
else:
return False
def DoesTree1HaveTree2(self, pRoot1, pRoot2):
"""递归比较,两颗子树的值"""
if pRoot2 == None:
return True
if pRoot1 == None:
return False
if not self.Equal(pRoot1.val, pRoot2.val):
return False
return self.DoesTree1HaveTree2(pRoot1.left, pRoot2.left) and \
self.DoesTree1HaveTree2(pRoot1.right, pRoot2.right)
def HasSubtree(self, pRoot1, pRoot2):
"""递归的进行先根序遍历,从根节点开始对值进行比较"""
result = False
if pRoot1 != None and pRoot2 != None:
if self.Equal(pRoot1.val, pRoot2.val):
result = self.DoesTree1HaveTree2(pRoot1, pRoot2)
if not result:
result = self.HasSubtree(pRoot1.left, pRoot2)
if not result:
result = self.HasSubtree(pRoot1.right, pRoot2)
return result
# test code
# 已在牛客网通过测试
# + [markdown] heading_collapsed=true
# # 面试题27:二叉树的镜像
# 题目:请完成一个函数,输入一颗二叉树,输出它的镜像。
# + hidden=true
class Solution:
def Mirror(self, root):
"""递归的交换二叉树的左右子树,遇叶子节点停止递归"""
if root == None:
return
if root.left == None and root.right == None:
return
pTemp = root.left
root.left = root.right
root.right = pTemp
if root.left:
self.Mirror(root.left)
if root.right:
self.Mirror(root.right)
# test code
# 已在牛客网通过测试
# + [markdown] heading_collapsed=true
# # 面试题28:对称的二叉树
# 题目:请实现一个函数,用来判断一颗二叉树是不是对称的。如果一颗二叉树和它的镜像一样,那么它是对称的。
# + hidden=true
class Solution:
def isSymmetricalCore(self, pRoot1, pRoot2):
"""递归比较前序遍历和对称前序遍历序列的值,若相同,则为对称二叉树"""
if pRoot1 == None and pRoot2 == None:
return True
if pRoot1 == None or pRoot2 == None:
return False
if pRoot1.val != pRoot2.val:
return False
return self.isSymmetricalCore(pRoot1.left,pRoot2.right) and self.isSymmetricalCore(pRoot1.right, pRoot2.left)
def isSymmetrical(self, pRoot):
"""调用核心函数,利用传参避免指向同一个对象"""
return self.isSymmetricalCore(pRoot, pRoot)
# test code
# 已在牛客网上通过测试
# + [markdown] heading_collapsed=true
# # 面试题29:顺时针打印矩阵
# 题目:输入一个举着,按照从外向里以顺时针的顺序依次打印出每一个数字。例如:
# [[1,2,3,4]
# [5,6,7,8]
# [9,10,11,12]
# [13,14,15,16]]
# 则依次打印出数字1,2,3,4,8,12,16,15,14,13,9,5,6,7,11,10。
# + hidden=true
class Solution:
def PrintMatrixInCircle(self, matrix, start, result):
"""根据给定的start打印二维矩阵的一圈,并将其存入result中,因为result是列表、可变类型,函数不返回
任何参数也可对传入的result中的元素进行修改"""
endX = len(matrix[0]) - 1 - start
endY = len(matrix) - 1 - start
#从左到右打印一行
for i in range(start, endX + 1):
number = matrix[start][i]
result.append(number)
#从上到下打印一列
if start < endY:
for i in range(start + 1, endY + 1):
number = matrix[i][endX]
result.append(number)
#从右到左打印一行
if start < endX and start < endY:
for i in range(endX - 1, start - 1, -1):
number = matrix[endY][i]
result.append(number)
#从下到上打印一列
if start < endX and start < endY - 1:
for i in range(endY - 1, start, -1):
number = matrix[i][start]
result.append(number)
def printMatrix(self, matrix):
"""循环打印每一圈矩阵"""
result = []
if not matrix or len(matrix) < 0 or len(matrix[0]) < 0:
return result
start = 0
while len(matrix[0]) > start*2 and len(matrix) > start*2:
self.PrintMatrixInCircle(matrix, start, result)
start += 1
return result
# test code
# 已在牛客网通过测试
# -
# # 面试题30:包含min函数的栈
# 题目:定义栈的数据结构,请在该类型中实现一个能够得到栈的最小元素的min函数。在该栈中,调用min、push、pop的时间复杂度都是O(1)。
# +
class Solution:
"""维护一个和data相同大小的辅助栈,用来存储最小值"""
def __init__(self):
self.m_data = []
self.m_min = []
def push(self, node):
"""压入数据的同时维护辅助栈,使的辅助栈中存储的最小值始终和数据栈中具有相同的位置"""
self.m_data.append(node)
if len(self.m_min) == 0 or node < self.m_min[-1]:
self.m_min.append(node)
else:
self.m_min.append(self.m_min[-1])
def pop(self):
if len(self.m_min) > 0 and len(self.m_data) > 0:
self.m_data.pop(-1)
self.m_min.pop(-1)
else:
assert Exception("data is empty")
def top(self):
return self.m_data[-1]
def min(self):
return self.m_min[-1]
# test code
# 已在牛客网通过测试
# + [markdown] heading_collapsed=true
# # 面试题31:栈的压入、弹出序列
# 题目:输入两个整数的序列,第一个序列表示栈的压入顺序,请判断第二个序列是否为该栈的弹出顺序。假设压入栈的所有数字均不相等。例如,序列[1,2,3,4,5]是某栈的压入序列,序列[4,5,3,2,1]是该压栈序列对应的一个弹出序列,但[4,3,5,1,2]就不可能是该压栈序列的弹出序列。
# + hidden=true
class Solution:
def IsPopOrder(self, pushV, popV):
"""利用辅助栈模拟栈的压入和弹出,与弹出序列比较若不符合则返回False"""
bPossible = False
if pushV != None and popV != None:
pNextPush = 0
pNextPop = 0
stackData = []
nLength = len(pushV)
while pNextPop < nLength:
while not stackData or stackData[-1] != popV[pNextPop]:
if pNextPush == nLength:
break
stackData.append(pushV[pNextPush])
pNextPush += 1
if stackData[-1] != popV[pNextPop]:
break
stackData.pop(-1)
pNextPop += 1
if not stackData and pNextPop == nLength:
bPossible = True
return bPossible
# test code
# 已在牛客网通过测试
# + [markdown] heading_collapsed=true
# # 面试题32:从上到下打印二叉树
# # + 题目一:不分行从上到下打印二叉树
# 从上到下打印出二叉树的每个节点,同一层的节点按照从左到右收尾顺序打印。
# + hidden=true
class Solution:
def PrintFromTopToBottom(self, root):
"""利用队列的先进先出原则,顺序存储要打印的数值"""
result = []
if not root:
return result
dequeTreeNode = []
dequeTreeNode.append(root)
while len(dequeTreeNode):
pNode = dequeTreeNode.pop(0)
result.append(pNode.val)
if pNode.left:
dequeTreeNode.append(pNode.left)
if pNode.right:
dequeTreeNode.append(pNode.right)
return result
# test code
# 已在牛客网通过测试
# + [markdown] hidden=true
# # + 题目二:分行从上到下打印二叉树
# 从上到下按层打印二叉树,同一层的节点按照从左到右的顺序打印,每一层打印一行。
# + hidden=true
class Solution:
# 返回二维列表[[1,2],[4,5]]
def Print(self, pRoot):
"""在利用队列的基础上,额外加两个变量表示下一行的节点数和当前行剩余位打印节点数"""
result = [[]]
if not pRoot:
return []
nodes = []
nodes.append(pRoot)
nextLevel = 0
toBePrinted = 1
while nodes:
pNode = nodes.pop(0)
result[-1].append(pNode.val)
if pNode.left:
nodes.append(pNode.left)
nextLevel += 1
if pNode.right:
nodes.append(pNode.right)
nextLevel += 1
toBePrinted -= 1
if toBePrinted == 0 and nextLevel != 0:
result.append([])
toBePrinted = nextLevel
nextLevel = 0
return result
# test code
# 已在牛客网通过测试
# + [markdown] hidden=true
# # + 题目三:之字形打印二叉树
# 请实现一个函数按照之字形顺序打印二叉树,即第一行按照从左到右的顺序打印,第二层按照从右到左的顺序打印,第三行再按照从左到右的顺序打印,其他行以此类推。
# + hidden=true
class Solution:
# 返回二维列表[[1,2],[4,5]]
def Print(self, pRoot):
"""利用栈的先进后出原则,使用两个栈分别存储当前行和下一行,在弹出元素的同时压入下一行元素到另一个栈中"""
if not pRoot:
return []
result = [[]]
levels = [[],[]]
current = 0
next = 1
levels[current].append(pRoot)
while levels[0] or levels[1]:
pNode = levels[current].pop()
result[-1].append(pNode.val)
if current == 0:
if pNode.left:
levels[next].append(pNode.left)
if pNode.right:
levels[next].append(pNode.right)
else:
if pNode.right:
levels[next].append(pNode.right)
if pNode.left:
levels[next].append(pNode.left)
if not levels[current] and levels[next]:
result.append([])
current = 1 - current
next = 1 - next
return result
# test code
# 代码已在牛客网通过测试
# + [markdown] heading_collapsed=true
# # 面试题33:二叉搜索树的后序遍历序列
# 题目:输入一个整数数组,判断该数组是不是某二叉搜索树的后序遍历结果。如果是则返回true,否则返回false。假设输入的数组的任意两个数字互不相同。
# + hidden=true
class Solution:
def VerifySquenceOfBST(self, sequence):
"""先提取末尾元素作为根节点,然后递归验证左右子树"""
if not sequence:
return False
root = sequence[-1]
i = 0
#使用while循环使的i总是指向第一个非左子树元素
while i < len(sequence) - 1:
if sequence[i] > root:
break
i += 1
for j in range(i, len(sequence) - 1):
if sequence[j] < root:
return False
left = True
if i > 0:
left = self.VerifySquenceOfBST(sequence[:i])
right = True
if i < len(sequence) - 1:
right = self.VerifySquenceOfBST(sequence[i:-1])
return left and right
# test code
# 代码已在牛客网测试通过
# + [markdown] heading_collapsed=true
# # 面试题34:二叉树中和为某一值的路径
# 题目:输入一颗二叉树和一个整数,打印出二叉树中节点值的和为输入整数的所有路径。从树的根节点开始往下一直到叶节点所经过的节点形成一条路径
# + hidden=true
class Solution:
# 返回二维列表,内部每个列表表示找到的路径
def FindPathCore(self,root, expectNumber, path, result):
"""递归遍历左子树、右子树,path列表记录一次遍历路径,result列表记录符合条件的路径,因path,result都是可变类型
故,函数中可不设返回值就能进行参数传递。另由于二叉树一般填满左子树再填充右子树,故先检查左子数,再检查右子树即可满足
牛客网中较长的路径在前,较短路径在后的要求,但此方法对一般二叉树不适用,还有改进空间"""
path.append(root.val)
if sum(path) == expectNumber and (root.left == None and root.right == None):
#找到一个匹配的路径
#使用path[:]的方式浅拷贝列表到result中,避免直接使用path(引用)造成后续path发生变化,引起result也改变。
#注:path中元素为基本常量类型时可采用此方法,否侧需要使用深拷贝
result.append(path[:])
if root.left != None:
self.FindPathCore(root.left,expectNumber,path, result)
if root.right != None:
self.FindPathCore(root.right,expectNumber,path,result)
path.pop()
def FindPath(self, root, expectNumber):
"""主函数,检查输入节点是否为空"""
if not root:
return []
path = []
result = []
self.FindPathCore(root, expectNumber,path, result)
return result
# test code
# 代码已在牛客网通过测试
# + [markdown] heading_collapsed=true
# # 面试题35:复杂链表的复制
# 题目:请实现函数,复制一个复杂链表。在复杂链表中,每个节点除了有一个next指针指向下一个节点,还有一个pSibling指针指向链表中的任意节点或者nullptr
# + hidden=true
class Solution:
# 返回 RandomListNode
def CloneNodes(self,phead):
"""在原有的链表基础上进行赋值,即将每个节点沿着next链复制一个"""
pNode = phead
while pNode != None:
pCloned = RandomListNode(None)
pCloned.label = pNode.label
pCloned.next = pNode.next
pCloned.random = None
pNode.next = pCloned
pNode = pCloned.next
def ConnetSiblingNodes(self, pHead):
"""修复复制后链表节点的random链接"""
pNode = pHead
while pNode != None:
pClone = pNode.next
if pNode.random != None:
pClone.random = pNode.random.next
pNode = pClone.next
def ReconnectNodes(self, pHead):
"""将复制好的链表拆分成两个链表,奇数位为原链表,偶数位为赋值好的链表"""
pNode = pHead
pCloneHead = None
pCloneNode = None
if pNode != None:
pCloneHead = pCloneNode = pNode.next
pNode.next = pCloneNode.next
pNode = pNode.next
while pNode != None:
pCloneNode.next = pNode.next
pCloneNode = pCloneNode.next
pNode.next = pCloneNode.next
pNode = pNode.next
return pCloneHead
def Clone(self, pHead):
"""在原链表基础上进行复制,再拆分,分成三个步骤,三个函数依次进行处理"""
self.CloneNodes(pHead)
self.ConnetSiblingNodes(pHead)
return self.ReconnectNodes(pHead)
# test code
# 代码已在牛客网通过测试
# + [markdown] heading_collapsed=true
# # 面试题36:二叉搜索树与双向链表
# 题目:输入一颗二叉搜索树,将该二叉搜索树转换成一个排序的双向链表。要求不能创建任何新的节点,只能调整树中节点指针的指向
# + hidden=true
class Solution:
def ConvertNode(self,pNode, pLastNodeInList):
"""递归的转换节点的左子树和右子树,返回转换成链表后的最后一个节点
注意:pLastNodeList对象本身在函数中是不可更改的,若强行修改相当于定义了同名的局部变量,因此必须用return的方式返回修改值"""
if pNode == None:
return
pCurrent = pNode
if pCurrent.left != None:
pLastNodeInList = self.ConvertNode(pCurrent.left, pLastNodeInList)
pCurrent.left = pLastNodeInList
if pLastNodeInList != None:
pLastNodeInList.right = pCurrent
pLastNodeInList = pCurrent
if pCurrent.right != None:
pLastNodeInList = self.ConvertNode(pCurrent.right, pLastNodeInList)
return pLastNodeInList
def Convert(self, pRootOfTree):
"""通过调用转换函数,并调整返回的末尾节点,返回头节点"""
pLastNodeInList = None
pLastNodeInList = self.ConvertNode(pRootOfTree,pLastNodeInList)
pHeadOfList = pLastNodeInList
while pHeadOfList != None and pHeadOfList.left != None:
pHeadOfList = pHeadOfList.left
return pHeadOfList
# + [markdown] heading_collapsed=true
# # 面试题37:序列化二叉树
# 题目:请实现两个函数,分别用来序列化和反序列化二叉树
# + hidden=true
class Solution:
def Serialize(self, root):
"""递归的前序遍历二叉树,遇空节点使用特殊字符代替"""
result = []
if root == None:
result.append(None)
return result
result.append(root.val)
result = result + (self.Serialize(root.left))
result = result + (self.Serialize(root.right))
return result
def Deserialize(self, s):
"""递归的读取序列生成节点"""
if not s:
return None
value = s.pop(0)
pRoot = None
if value:
pRoot = TreeNode(value)
pRoot.left = self.Deserialize(s)
pRoot.right = self.Deserialize(s)
return pRoot
# + [markdown] heading_collapsed=true
# # 面试题38:字符串的排列
# 题目:输入一个字符串,打印出该字符串中字符的所有排列。例如,输入字符abc,则打印出由字符a,b,c所能排列出来的所有字符串abc,acb,bac,bca,cab,cba。
# + hidden=true
class Solution:
"""依照剑指offer中C++代码思路修改的python算法,原C++代码中无法保证输入重复字符时,输出不重复的组合,也无法保证按字典序输出。
本算法中在循环中添加if判断是否字符是否重复,并在Permutation函数返回时,调用sorted函数进行排序,因此有很大的算法改进空间"""
def PermutationCore(self, ss, i):
"""递归的从左到右调换列表中的元素位置,返回调换后的组合,i为调换的起始位置"""
result = []
if i >= len(ss) - 1:
# 当起始位置到达列表末尾时,直接返回该列表组合成的字符串即可
result.append(''.join(ss))
return result
else:
for j in range(i, len(ss)):
# 循环中依次将每个字符和起始字符交换,递归返回剩余部分组合的字符串
if ss[j] == ss[i] and j != i:
# 若两字符相同,则没有交换的必要(首字符交换的情况除外)
continue
temp = ss[j]
ss[j] = ss[i]
ss[i] = temp
result = result + self.PermutationCore(ss, i+1)
# 还原交换过的字符
temp = ss[j]
ss[j] = ss[i]
ss[i] = temp
return result
def Permutation(self, ss):
"""主函数,传入列表,使用sorted函数按字典序重新排序返回的字符串组合"""
if not ss:
return ''
return sorted(self.PermutationCore(list(ss), 0))
# test code
# 已在牛客网通过测试
# + [markdown] heading_collapsed=true
# # 面试题39:数组中出现次数超过一半的数字
# 题目:数组中有一个数字出现的次数超过数组长度的一半,请找出这个数字。例如,输入一个长度为9的数组[1,2,3,2,2,2,5,4,2]。由于数组2在数组中出现了5次,超过数组长度的一半,因此输出2.
# + code_folding=[] hidden=true
class Solution:
def CheckMoreThanHalf(self, numbers , number):
"""检查输入数字重复次数是否超过数组长度的一半"""
times = 0
for i in range(len(numbers)):
if numbers[i] == number:
times += 1
isMoreThanHalf = True
if times *2 <= len(numbers):
isMoreThanHalf = False
return isMoreThanHalf
def MoreThanHalfNum_Solution(self, numbers):
"""通过加减法统计数字在数组中出现的次数,得到次数最高的数字,最终判断其次数是否超过数组长度的一半,算法复杂度O(n)"""
if not numbers:
return 0
result = numbers[0]
times = 1
for i in range(len(numbers)):
if times == 0:
result = numbers[i]
times = 1
elif numbers[i] == result:
times += 1
else:
times -= 1
if not self.CheckMoreThanHalf(numbers, result):
result = 0
return result
# test code
# 已在牛客网通过测试
# + code_folding=[2] hidden=true
# 另一个思路,利用快速排序法的思想,(此代码尚存在bug通过牛客网测试)
class Solution:
def __init__(self):
"""初始化全局错误指针,为了能使用random标准库,用self.random去应用导入后的该库"""
self.g_bInputInvalid = False
import random
self.random = random
def Partition(self, data, start, end):
"""随机选择基准数,将较小数移到前方,返回基准数在调整顺序后的索引位置"""
if not data:
raise Exception("Invalid Parameters")
# 在快速排序法基础上额外添加的防止错误出入代码
if start == end:
return start
index = self.random.randrange(start, end)
temp = data[index]
data[index] = data[end]
data[end] = temp
small = start - 1
for index in range(start, end):
if data[index] < data[end]:
small += 1
if small != index:
temp = data[index]
data[index] = data[small]
data[small] = temp
small += 1
temp = data[small]
data[small] = data[end]
data[end] = temp
return small
def CheckMoreThanHalf(self, numbers, number):
"""同上一cell方法的检查函数"""
times = 0
for i in range(len(numbers)):
if numbers[i] == number:
times += 1
isMoreThanHalf = True
if times * 2 <= len(numbers):
isMoreThanHalf = False
self.g_bInputInvalid = True
return isMoreThanHalf
def CheckInvalidArray(self, numbers):
"""检查输入是否合法"""
self.g_bInputInvalid = False
if not numbers:
self.g_bInputInvalid = True
return self.g_bInputInvalid
def MoreThanHalfNum_Solution(self, numbers):
"""基于快速排序法,重复次数超过数组长度一半时该元素必定位于排序后数组的中间位置,因此基于快速排序法查找排序后位于中间位置
的元素,并检查该元素是否符合要求"""
if self.CheckInvalidArray(numbers):
return 0
middle = len(numbers) // 2
start = 0
end = len(numbers) - 1
index = self.Partition(numbers,start, end)
while index != middle:
if index > middle:
end = index - 1
index = self.Partition(numbers,start, end)
else:
start = index + 1
index = self.Partition(numbers, start, end)
result = numbers[middle]
if not self.CheckMoreThanHalf(numbers, result):
result = 0
return result
# + [markdown] heading_collapsed=true
# # 面试题40:最小的k个数
# 题目:输入n个整数,找出其中最小的k个数。例如,输入4,5,1,6,2,7,3,8这8个数,则最小的四个数是1,2,3,4.
# + code_folding=[3] hidden=true
class Solution:
"""时间复杂度为O(n)的算法,需要修改数组,且输出的k个数不一定是排序的,此算法虽能满足题目要求,但不能通过牛客网测试"""
def Partition(self, data, start, end):
"""随机选择基准数,将较小数移到前方,返回基准数在调整顺序后的索引位置"""
if not data:
raise Exception("Invalid Parameters")
# 在快速排序法基础上额外添加的防止错误出入代码
if start == end:
return start
index = random.randrange(start, end)
temp = data[index]
data[index] = data[end]
data[end] = temp
small = start - 1
for index in range(start, end):
if data[index] < data[end]:
small += 1
if small != index:
temp = data[index]
data[index] = data[small]
data[small] = temp
small += 1
temp = data[small]
data[small] = data[end]
data[end] = temp
return small
def GetLeastNumbers_Solution(self, tinput, k):
if not tinput or k <= 0:
return
start = 0
end = len(tinput) - 1
index = self.Partition(tinput, start, end)
while index != k - 1:
if index > k - 1:
end = index - 1
index = self.Partition(tinput, start, end)
else:
start = index + 1
index = self.Partition(tinput, start, end)
return tinput[:k]
#test code
a = Solution()
print(a.GetLeastNumbers_Solution([4,5,1,6,2,7,3,8], 1))
# + hidden=true
class Solution:
"""解法二:时间复杂度为O(nlogK)的算法,利用红黑树来维护最大值,适合用来处理海量数据"""
def GetLeastNumbers_Solution(self, tinput, k):
if not tinput or len(tinput) < k or k <= 0:
return []
result = []
for i in tinput:
if len(result) < k:
result.append(i)
else:
result.sort()
if i < result[-1]:
result[-1] = i
return sorted(result)
# -
# # 面试题41:数据流中的中位数
# 题目:如何得到一个数据流中的中位数?如果从数据流中独处奇数个数值,那么中位数就是所有数值排序之后位于中间的数值,若果从数据流中读出偶数个数值,那么中位数就是所有数值排序后中间两个数的平均值
class Solution:
"""使用最大堆和最小堆来存储排好序后的中位数左边和右边,根据两堆的总元素两即可判断中位数的数值"""
def __init__(self):
"""利用对象的初始化功能,导入heapq优先队列堆标准库,并初始化最大堆和最小堆"""
import heapq
self.hq = heapq
self.max_hq = []
self.min_hq = []
def Insert(self, num):
"""数据流的个数是偶数个则在最小堆中插入,同时判断新输入的数据流插入后是否符合最大堆的最大元素小于最小堆的最小元素。
由于heapq标准库中只有最小堆,所以采用所有数据取反存储的方式实现最大堆"""
if ((len(self.min_hq) + len(self.max_hq)) & 1) == 0:
if len(self.max_hq) > 0 and num < -self.max_hq[0]:
self.hq.heappush(self.max_hq, -num)
num = -self.hq.heappop(self.max_hq)
self.hq.heappush(self.min_hq, num)
else:
if len(self.min_hq) > 0 and self.min_hq[0] < num:
self.hq.heappush(self.min_hq, num)
num = self.hq.heappop(self.min_hq)
self.hq.heappush(self.max_hq, -num)
def GetMedian(self, fuck):
"""判断数据流的大小,按照奇偶规则从堆中返回中位数,另外由于牛客网bug,此函数必须额外增加一个参数,遂用fuck代替"""
size = len(self.max_hq) + len(self.min_hq)
if size == 0:
raise Exception("No number are available!")
median = 0
if (size & 1) == 1:
median = self.min_hq[0]
else:
median = float(self.min_hq[0] - self.max_hq[0]) / 2
return median
# test code
# 已在牛客网通过测试
# + [markdown] heading_collapsed=true
# # 面试题42:连续子数组的最大和
# 题目:输入一个整形数组,数组中有正数也有负数。数组中的一个或连续多个整数组成一个子数组。求所有子数组的和的最大值。要求时间复杂度为O(n)
# + hidden=true
class Solution:
def FindGreatestSumOfSubArray(self, array):
"""一次遍历,逐个比较累加子数组之和,若当前和小于等于0则重新选择下一值为新起点"""
if not array:
return False
nCurSum = 0
nGreatSum = -0x80000000 # python中数值溢出会自动使用大数表示法,因此要表示最小整型数字只能在前方加‘-’号
for i in array:
if nCurSum <= 0:
nCurSum = i
else:
nCurSum += i
if nCurSum > nGreatSum:
nGreatSum = nCurSum
return nGreatSum
# test code
# 代码已在牛客网通过测试
# + [markdown] heading_collapsed=true
# # 面试题43:[1,n]整数中1出现的次数
# 题目:输入一个整数n,求[1,n]这n个整数的十进制表示中1出现的次数。例如,输入12,则[1,12]这些整数中包含1的数字有1,10,11和12,1一共出现了5次
# + hidden=true
class Solution:
def NumberOf1(self,strN):
"""递归的计算含有1的数量"""
if not strN:
return 0
first = ord(strN[0]) - ord('0')
length = len(strN)
if length == 1 and first == 0:
return 0
if length == 1 and first > 0:
return 1
numFirstDigit = 0
if first > 1:
numFirstDigit = pow(10, length - 1)
elif first == 1:
numFirstDigit = int(''.join(strN[1:])) + 1
numOtherDigts = first*(length - 1)*pow(10, length - 2)
numRecursive = self.NumberOf1(strN[1:])
return numFirstDigit + numOtherDigts + numRecursive
def NumberOf1Between1AndN_Solution(self, n):
"""主函数,判断输入数字是否有效,并将其转换成字符型列表,算法复杂度O(logn)"""
if n <= 0:
return 0
return self.NumberOf1(list(str(n)))
# test code
# 代码已在牛客网通过测试
# + [markdown] heading_collapsed=true
# # 面试题44:数字序列中某一位的数字
# 题目:数字以0123456789101112131415...的格式序列化到一个字符序列中。在这个序列中,第5位(从0开始计数)是5,第13位是1,第19为是4,等等。请写一个函数,求任意第n位对应的数字
# + hidden=true
class Solution:
def beginNumber(self, digits):
if digits == 1:
return 0
return pow(10, digits - 1)
def digitsAtIndex(self,index, digits):
number = self.beginNumber(digits) + index / digits
indexFromRight = digits - index % digits
for i in range(1,indexFromRight):
number /= 10
return int(number % 10)
def countOfIntegers(self, digits):
if digits == 1:
return 10
count = pow(10, digits - 1)
return 9*count
def digitsAtIndex_main(self, index):
if index < 0:
return -1
digits = 1
while True:
number = self.countOfIntegers(digits)
if index < number * digits:
return self.digitsAtIndex(index, digits)
index -= digits * number
digits += 1
return -1
# test code
a = Solution()
data = 10
print(a.digitsAtIndex_main(data))
# + [markdown] heading_collapsed=true
# # 面试题45:把数组排成最小的数
# 题目:输入一个正整数数组,把数组里所有数字拼接起来排成一个数,打印能拼接出的所有数组中最小的一个。例如,输入数组{3,32,321},则打印出这三个数字能排成的最小数字321323
# + hidden=true
class Solution:
"""定义一种新的比较大小数字大小的方法,使用该方法对数组进行排序,同时考虑大数问题"""
def PrintMinNumber(self, numbers):
if not numbers:
return ''
strNumber = [str(i) for i in numbers]
def compare(str1, str2):
"""新的比较方法,尝试拼接两字符串然后比较大小"""
if str1+str2 > str2+str1:
return 1
elif str1 + str2 < str2 + str1:
return -1
else:
return 0
# python3中取消了cmp参数,但牛客网中使用的是python2.7故能通过测试代码,python3中的解决代码如下;
# import functools
# strNumber = sorted(strNumber, key=functools.cmp_to_key(compare))
strNumber = sorted(strNumber, cmp=lambda x,y:compare(x, y))
return ''.join(strNumber)
# test code
# 已在牛客网通过测试
# + [markdown] heading_collapsed=true
# # 面试题46:把数字翻译成字符串
# 题目:给定一个数字,我们按照如下规则把它翻译为字符串:0翻译成'a',1翻译为'b',....,11翻译为'l',...,25翻译为'z'。一个数字可能有多个翻译。例如,12258有5中不同的翻译,分别是'bccfi','bwfi','bczi','mcfi','mzi'。请编程实现一个函数,用来计算一个数字有多少种不同的翻译方法。
# + hidden=true
class Solution:
"""从右向左的依次计算可组合翻译的种类,避免重复计算了拆分的小问题"""
def GetTranslationCount(self,number):
if number < 0:
return 0
strNumber = str(number)
length = len(strNumber)
counts = [None] * length
count = 0
for i in range(length - 1, -1, -1):
count = 0
if i < length - 1: # 避免counts在未初始化时对count进行了赋值
count = counts[i+1] # count初始值总是前面可组合的次数,在此基础上进行累积
else:
count = 1
if i < length - 1:
digit1 = ord(strNumber[i]) - ord('0')
digit2 = ord(strNumber[i+1]) - ord('0')
converted = digit1 * 10 + digit2
if converted >= 10 and converted <= 25: # 可合并翻译的情况
if i < length - 2:
count += counts[i+2]
else:
count += 1
counts[i] = count
return count
# test code
a = Solution()
data = 12258
print(a.GetTranslationCount(data))
# + [markdown] heading_collapsed=true
# # 面试题47:礼物的最大价值
# 题目:在一个m*n的棋盘的每一格都放有一个礼物,每个礼物都有一定的价值(价值大于0).你可以从棋盘的左上角开始拿格子里的礼物,并每次向右或者向下移动一格,直到达到棋盘的右下角。给定一个棋盘及其上面的礼物,请计算你最多能拿到多少价值的礼物?
# + hidden=true
class Solution:
def getMaxValue_solution(self,values):
"""解决方案一,使用辅助的二维矩阵记录路径上能得到的最大价值"""
if not values:
return 0
rows = len(values)
clos = len(values[0])
maxValue = [[None] * clos for _ in range(rows)]
for i in range(rows):
for j in range(clos):
left = 0
up = 0
if i > 0:
up = maxValue[i-1][j]
if j > 0:
left = maxValue[i][j-1]
maxValue[i][j] = max(left, up) + values[i][j]
maxValue = maxValue[rows-1][clos-1]
return maxValue
def getMaxValue_solution2(self,values):
"""优化方案,使用一维数组记录路径上能得到的最大价值"""
if not values:
return 0
rows = len(values)
clos = len(values[0])
maxValues = [None] * clos
for i in range(rows):
for j in range(clos):
left = 0
up = 0
if i > 0 :
up = maxValues[j]
if j > 0:
left = maxValues[j-1]
maxValues[j] = max(up, left) + values[i][j]
maxValues = maxValues[clos - 1]
return maxValues
# test code
a = Solution()
data = [[1,10,3,8],[12,2,9,6],[5,7,4,11],[3,7,16,5]]
print(a.getMaxValue_solution2(data))
# + [markdown] heading_collapsed=true
# # 面试题48:最长不含重复字符的字符串
# 题目:请从字符串中找出一个最长的不包含重复字符的子字符串,计算该最长子字符串的长度。假设字符串中只包含'a'-'z'的字符。例如,在字符串"arabcacfr"中,最长的不含重复字符的子字符串是"acfr",长度为4.
# + hidden=true
class Solution:
def longestSubstringWithoutDuplication(self, strs):
"""position用来存储上一次对应位置字符出现的索引值"""
curLength = 0
maxLength = 0
position = [-1] * 26
for i in range(len(strs)):
prevIndex = position[ord(strs[i]) - ord('a')]
if prevIndex < 0 or i - prevIndex > curLength: # 计算距离d是否大于当前长度
curLength += 1
else:
if curLength > maxLength:
maxLength = curLength
curLength = i - prevIndex
position[ord(strs[i]) - ord('a')] = i
if curLength > maxLength:
maxLength = curLength
return maxLength
# test code
a = Solution()
data = 'arabcacfr'
print(a.longestSubstringWithoutDuplication(data))
# + [markdown] heading_collapsed=true
# # 面试题49:丑数
# 题目:我们把只包含因子2,3,5的数成为丑数,求从小到大的顺序的第1500个丑数。例如,6,8都是丑数,但14不是,因为它包含因子7.习惯上我们把1当做第一个丑数。
# + hidden=true
class Solution:
def GetUglyNumber_Solution(self, index):
"""利用丑数产生丑数,相比于直接判断每一个整数的方法计算速度要快,但同时将使用O(n)复杂度的存储空间"""
if index <= 0:
return 0
pUglyNumbers = [None] * index
pUglyNumbers[0] = 1
nextUglyIndex = 1
# pMultiply's Index in pUglyNumbers 记录第一个乘以因子比当前最大丑数大的基丑数的索引位置
pMultiply2 = 0
pMultiply3 = 0
pMultiply5 = 0
while nextUglyIndex < index:
minNumber = min([pUglyNumbers[pMultiply2] * 2, pUglyNumbers[pMultiply3] * 3,
pUglyNumbers[pMultiply5] * 5])
pUglyNumbers[nextUglyIndex] = minNumber
# update T2,T3,T5
while pUglyNumbers[pMultiply2] * 2 <= pUglyNumbers[nextUglyIndex]:
pMultiply2 += 1
while pUglyNumbers[pMultiply3] * 3 <= pUglyNumbers[nextUglyIndex]:
pMultiply3 += 1
while pUglyNumbers[pMultiply5] * 5 <= pUglyNumbers[nextUglyIndex]:
pMultiply5 += 1
nextUglyIndex += 1
ugly = pUglyNumbers[nextUglyIndex - 1]
return ugly
# test code
# 代码已在牛客网通过测试
# -
# # 面试题50:第一个只出现一次的字符
# + 题目一:字符串中第一个只出现一次的字符。
# 如输入"abaccdeff"则输出'b'
class Solution:
def FirstNotRepeatingChar(self, s):
"""使用长度为256的列表表示ASCII码,列表中的值表示对应字符出现的次数,模拟哈希表,统计字符出现的次数,
再依照字符出现的顺序检查哈希表,找出第一次只出现一次的字符"""
if not s:
return ''
tableSize = 256
hashTable = [0] * tableSize
for i in range(len(s)):
hashTable[ord(s[i])] += 1
for i in range(len(s)):
if hashTable[ord(s[i])] == 1:
return s[i]
return ''
# test code
a = Solution()
data = 'abaccdeff'
print(a.FirstNotRepeatingChar(data))
# 牛客网对应代码
"""在一个字符串(0<=字符串长度<=10000,全部由字母组成)中找到第一个只出现一次的字符,并返回它的位置,
如果没有则返回 -1(需要区分大小写)"""
class Solution:
def FirstNotRepeatingChar(self, s):
"""使用长度为256的列表表示ASCII码,列表中的值表示对应字符出现的次数,模拟哈希表,统计字符出现的次数,
再依照字符出现的顺序检查哈希表,找出第一次只出现一次的字符,并在第一题的基础上返回字符改为范围该索引,
并将返回空字符的代码改为返回-1"""
if not s:
return -1
tableSize = 256
hashTable = [0] * tableSize
for i in range(len(s)):
hashTable[ord(s[i])] += 1
for i in range(len(s)):
if hashTable[ord(s[i])] == 1:
return i
return -1
# test code
# 代码已在牛客网测试通过
# + 题目二:字符流中第一个只出现一次的字符
# 请实现一个函数,用来找出字符流中的第一个只出现一次的字符。例如,当从字符流中只读出两个字符"go"时,第一个只出现一次的字符是"g";当从该字符流中读出前6个字符"google"时,第一个只出现一次的字符是"I"。
class Solution:
def __init__(self):
"""初始化哈希表和索引,其中
occurrence[i] = -1 表示元素尚未找到
occurrence[i] = -2 表示元素被找到多次
occurrence[i] >= 0 表示元素仅被找到一次"""
self.occurrence = [-1] * 256
self.index = 0
def Insert(self,ch):
if self.occurrence[ord(ch)] == -1:
self.occurrence[ord(ch)] = self.index
elif self.occurrence[ord(ch)] >= 0:
self.occurrence[ord(ch)] = -2
self.index += 1
def FirstAppearingOnce(self):
"""若元素不存在,或未被找到则输出#"""
ch = '#'
minIndex = 0xffffffff
for i in range(256):
if self.occurrence[i] >= 0 and self.occurrence[i] < minIndex:
ch = chr(i)
minIndex = self.occurrence[i]
return ch
# test code
# 代码已在牛客网通过测试
a = Solution()
data = 'abaccdebff'
for i in range(len(data)):
a.Insert(data[i])
print(a.FirstAppearingOnce())
# 存储数据版代码,上述算法并未在插入时存储有效的插入数据,此版本利用存储的数据对字符进行索引
class Solution:
def __init__(self):
"""初始化数据存储列表,散列表,和索引值"""
self.data = []
self.countNumber = [0]*256
self.once = 0
def FirstAppearingOnce(self):
"""利用始终指向第一个只出现一次的字符的索引值返回对应字符"""
return self.data[self.once] if self.once < len(self.data) else '#'
def Insert(self, char):
"""存储插入的字符并更新散列表中的次数,检查索引值是否始终指向第一次只出现一次的字符,若此字符不存在,则索引值指向界外"""
self.data.append(char)
self.countNumber[ord(char)] += 1
while self.once < len(self.data) and self.countNumber[ord(self.data[self.once])] != 1:
self.once += 1
# test code
# 代码已在牛客网通过测试
# + [markdown] heading_collapsed=true
# # 面试题51:数组中的逆序对
# 题目:在数组中有两个数字,如果前面一个数字大于后面的数字,则两个数字组成一个逆序对。输入一个数组,求出这个数组中的逆序对的总数。例如在数组{7,5,6,4}中,一共存在5个逆序对,分别是(7,6),(7,5),(7,4),(6,4),(5,4).
# + code_folding=[] hidden=true
class Solution:
def InverseParisCroe(self,data, copy, start, end):
"""利用归并排序原理,递归的将数组进行拆分,在拆分的内部进行比较计算逆序对数,再比较分组之间的大小和逆序对数合并分组"""
if start == end:
copy[start] = data[start]
return 0
length = (end - start) // 2
left = self.InverseParisCroe(copy,data,start,start + length)
right = self.InverseParisCroe(copy,data, start + length + 1, end)
i = start + length
j = end
indexCopy = end
count = 0
while i >= start and j >= start + length + 1:
if data[i] > data[j]:
copy[indexCopy] = data[i]
indexCopy -= 1
i -= 1
count += j - start - length
else:
copy[indexCopy] = data[j]
indexCopy -= 1
j -= 1
while i >= start:
copy[indexCopy] = data[i]
indexCopy -= 1
i -= 1
while j >= start + length + 1:
copy[indexCopy] = data[j]
indexCopy -= 1
j -= 1
return left + right + count
def InversePairs(self, data):
if not data:
return 0
copy = data[:]
count = self.InverseParisCroe(data, copy, 0, len(data) - 1)
return count
# test code
a = Solution()
data = [1,2,3,4,5,6,7,0]
print(a.InversePairs(data))
# + hidden=true
class Solution:
"""由于牛客网的原因,上个cell中的归并排序代码在算法复杂度上无法通过测试,但同样使用归并排序的以下算法可以
一下算法和上述归并排序原理的算法的区别在于,该算法在数组上从左到右的进行比较和统计逆序对数"""
def InversePairs(self, data):
return self.sort(data[:], 0, len(data)-1, data[:]) % 1000000007
def sort(self, temp, left, right, data):
if right - left < 1:
return 0
if right - left == 1:
if data[left] < data[right]:
return 0
else:
temp[left], temp[right] = data[right], data[left]
return 1
mid = (left + right) // 2
res = self.sort(data, left, mid, temp) + self.sort(data, mid+1, right, temp)
# 合并
i = left
j = mid + 1
index = left
while i <= mid and j <= right:
if data[i] <= data[j]:
temp[index] = data[i]
i += 1
else:
temp[index] = data[j]
res += mid - i + 1
j += 1
index += 1
while i <= mid:
temp[index] = data[i]
i += 1
index += 1
while j <= right:
temp[index] = data[j]
j += 1
index += 1
return res
# test code
# 已在牛客网通过测试
# + [markdown] heading_collapsed=true
# # 面试题52:两个链表的第一个公共节点
# 题目:输入两个链表,找出它们的第一个公共节点。
# + hidden=true
class Solution:
"""由于是单向链表,公共节点及其后的节点一定都相同,不会出现分叉。基于此算法先求出两链表的长度,让较长的链表先遍历多出的节点,
然后两链表再一起遍历,通过比较找出公共节点,若无公共节点返回None"""
def GetListLength(self,phaed):
nLength = 0
pNode = phaed
while pNode != None:
nLength += 1
pNode = pNode.next
return nLength
def FindFirstCommonNode(self, pHead1, pHead2):
nLength1 = self.GetListLength(pHead1)
nLength2 = self.GetListLength(pHead2)
nLengthDif = nLength1 - nLength2
pListHeadLong = pHead1
pListHeadShort = pHead2
if nLength2 > nLength1:
pListHeadLong = pHead2
pListHeadShort = pHead1
nLengthDif = nLength2 - nLength1
for i in range(nLengthDif):
pListHeadLong = pListHeadLong.next
while pListHeadLong != None and pListHeadShort != None and pListHeadLong != pListHeadShort:
pListHeadLong = pListHeadLong.next
pListHeadShort = pListHeadShort.next
return pListHeadLong
# test code
# 代码已在牛客网通过测试
# + [markdown] heading_collapsed=true
# # 面试题53:在排序数组中查找数字
# # + 题目一:数字在排序数字中出现的次数
# 统计一个数字在排序数组中出现的次数。例如,输入排序数组{1,2,3,3,3,3,4,5}和数字3,由于3在这个数组中出现了4次,因此输出4
# + hidden=true
class Solution:
def GetFirstK(self,data, k, start, end):
"""利用二分查找法原理查找第一个k出现的位置,二分法找到k之后再尝试判断前一位数值,用与判断当前找到的k是否是第一个k"""
if start > end:
return -1
middleIndex = (start + end) // 2
middleData = data[middleIndex]
if middleData == k:
if (middleIndex > 0 and data[middleIndex - 1] != k) or middleIndex == 0:
return middleIndex
else:
end = middleIndex - 1
elif middleData > k:
end = middleIndex - 1
else:
start = middleIndex + 1
return self.GetFirstK(data, k, start, end)
def GetLastK(self, data, k, start, end):
"""利用二分查找法原理查找最后一个k出现的位置,二分法找到k之后再尝试判断后一位数值,用与判断当前找到的k是否是最后一个k"""
if start > end:
return -1
middleIndex = (start + end) // 2
middleData = data[middleIndex]
if middleData == k:
if (middleIndex < len(data) - 1 and data[middleIndex + 1] != k) or middleIndex == len(data) - 1:
return middleIndex
else:
start = middleIndex + 1
elif middleData < k:
start = middleIndex + 1
else:
end = middleIndex - 1
return self.GetLastK(data, k, start, end)
def GetNumberOfK(self, data, k):
"""主函数,调用二分法原理的子函数,找到第一个k和最后一个k出现的位置,从而计算出k在数组中出现的次数"""
number = 0
if data:
first = self.GetFirstK(data, k, 0, len(data) - 1)
last = self.GetLastK(data, k, 0, len(data) - 1)
if first > -1 and last > -1:
number = last - first + 1
return number
# + [markdown] hidden=true
# # + 题目二:[0,n-1]中缺失的数字
# 一个长度为n-1的递增排序数组中的所有数字都是唯一的,并且每一个数字都在范围[0,n-1]之内。在范围[0,n-1]之内的n个数字有且只有一个数字不在该数组中,请找出该数字。
# + hidden=true
class Solution:
def GetMissingNumber(self,numbers):
"""问题转换为找到第一个下标和值不等的元素,利用二分查找法原理对排序数组进行查找"""
if not numbers:
return -1
left = 0
right = len(numbers) - 1
while left <= right:
middle = (right + left) >> 1
if numbers[middle] != middle:
if middle == 0 or numbers[middle-1] == middle - 1:
return middle
right = middle - 1
else:
left = middle + 1
if left == len(numbers):
return len(numbers)
return -1
# test code
a = Solution()
data = [0,1,2,4,5]
print(a.GetMissingNumber(data))
# + [markdown] hidden=true
# # + 题目三:数组中数值和下标相等的元素
# 假设一个单调递增的数组里每个元素都是整数并且唯一。请实现一个函数,找出数组中任意一个数值等于其下标的元素。例如,在数组{-3,-1,1,3,5}中,数字3和它的下标相等。
# + hidden=true
class Solution:
def GetNumberSameAsIndex(self,numbers):
if not numbers:
return -1
length = len(numbers)
left = 0
right = length - 1
while left <= right:
middle = (left + right) >> 1
if numbers[middle] == middle:
return middle
if numbers[middle] > middle:
right = middle - 1
else:
left = middle + 1
return -1
# test code
a = Solution()
data = [-3,-3,1,3,5]
print(a.GetNumberSameAsIndex(data))
# + [markdown] heading_collapsed=true
# # 面试题54:二叉搜索树的第K大节点
# 题目:给定一个二叉搜索数,请找出其中第k大的节点。例如,在下图中的二叉搜索树里,按节点数值大小顺序,第3大节点的值是4
# 
# + hidden=true
class TreeNode:
def __init__(self, x):
self.val = x
self.left = None
self.right = None
class Solution:
"""中根序遍历一遍二叉搜索树相当于按节点数值大小顺遍历二叉树,遍历k次即可找到第k大的节点"""
def KthNodeCroe(self,pRoot, k):
target = None
if pRoot.left != None:
target = self.KthNodeCroe(pRoot.left, k)
if target == None:
if k[0] == 1:
target = pRoot
k[0] -= 1
if target == None and pRoot.right != None:
target = self.KthNodeCroe(pRoot.right, k)
return target
def KthNode(self,pRoot, k):
if pRoot == None or k == 0:
return None
return self.KthNodeCroe(pRoot, [k])
# test code
# 代码已在牛客网通过测试
pRoot = TreeNode(5)
pRoot.left = TreeNode(3)
pRoot.right = TreeNode(7)
pRoot.left.left = TreeNode(2)
pRoot.left.right = TreeNode(4)
pRoot.right.left = TreeNode(6)
pRoot.right.right = TreeNode(8)
a = Solution()
print(a.KthNode(pRoot, 7).val)
# + [markdown] heading_collapsed=true
# # 面试题55:二叉树的深度
# # + 题目一:二叉树的深度
# 输入一颗二叉树的根节点,求该树的深度。从根节点到叶节点依次经过的节点(含根、叶节点)形成树的一条路径,最长路径的长度为树的深度
# + hidden=true
class Solution:
def TreeDepth(self, pRoot):
"""递归的计算左右子树的深度,其中较大者加1就是本树的深度"""
if pRoot == None:
return 0
nLeft = self.TreeDepth(pRoot.left)
nRight = self.TreeDepth(pRoot.right)
return nLeft + 1 if nLeft > nRight else nRight + 1
# test code
# 代码已在牛客网通过测试
# + [markdown] hidden=true
# # + 题目二:平衡二叉树
# 输入一颗二叉树的根节点,判断该树是不是平衡二叉树。如果某二叉树中任意节点的左、右子树的深度相差不超过1,那么它就是一颗平衡二叉树。
# + hidden=true
class Solution:
"""利用后根序遍历原理,遍历的同时比较两颗子树的深度,判断是否平衡,利用可变类型list传入递归函数取得被递归的左右子树的最大深度"""
def IsBalancedCroe(self, pRoot, pDepth):
if not pRoot:
pDepth[0] = 0
return True
left, right = [None], [None]
if self.IsBalancedCroe(pRoot.left, left) and self.IsBalancedCroe(pRoot.right, right):
diff = left[0] - right[0]
if diff <= 1 and diff >= -1:
pDepth[0] = 1 + (left[0] if left[0] > right[0] else right[0])
return True
return False
def IsBalanced_Solution(self, pRoot):
depth = [None]
return self.IsBalancedCroe(pRoot, depth)
# test code
# 代码已通过牛客网测试
# + [markdown] heading_collapsed=true
# # 面试题56:数组中数字出现的次数
# # + 题目-:数组中只出现一次的两个数字
# 一个整型数组里除了两个数字之外,其他数字都出现了两次。请写程序找出这两个只出现一次的数字。要求时间复杂度O(n),空间复杂度O(1)。
# + hidden=true
class Solution:
def IsBit1(self,num,indexBit):
"""判断num的从右到左第indexBit位是否是1"""
num = num >> indexBit
return (num & 1)
def FindFirstBitIs1(self,num):
"""检查num二进制中1位于从左到右的第几位,并返回位数"""
indexBit = 0
while (num & 1 == 0) and indexBit < 8*4:
num = num >> 1
indexBit += 1
return indexBit
def FindNumsAppearOnce(self, array):
"""用0作为初始值,从左到右连续对array每个元素进行异或,出现两次的元素最终会相互抵消,剩下的就是两个只出现一次的元素的
异或的结果resultExclusiveOR,调用函数获取结果二进制中最低位的位置indexOf1,并再次连续异或利用该位置对另一个只出现
一次的元素进行排除,从而获得的两个结果就是array中两个只出现一次的元素"""
if not array:
return
resultExclusiveOr = 0
for i in array:
resultExclusiveOr = resultExclusiveOr ^ i
indexOf1 = self.FindFirstBitIs1(resultExclusiveOr)
result = [0,0]
for i in array:
if self.IsBit1(i,indexOf1):
result[0] = result[0] ^ i
else:
result[1] = result[1] ^ i
return result
# test code
# 代码已在牛客网通过测试
# + [markdown] hidden=true
# # + 题目二:数组中唯一只出现一次的数字
# 在一个数组中除一个数字只出现一次之外,其他数字都出现了三次,请找出那个只出现一次的数字
# + hidden=true
class Solution:
def FindNumsAppearOnce(self, numbers):
"""将数组中所有元素的二进制数的每一位相加,若能被3整除则只出现一次的数字在此位一定为0,否则为1,收集这些1即可得到该数"""
if not numbers:
return None
bitSum = [0]*32
for i in range(len(numbers)):
bitMask = 1
for j in range(31,-1,-1):
bit = numbers[i] & bitMask
if bit != 0:
bitSum[j] += 1
bitMask = bitMask << 1
result = 0
for i in range(32):
result = result << 1
result += bitSum[i] % 3
return result
# test code
a = Solution()
data = [2,4,3,6,3,2,5,5,2,5,3,4,4]
print(a.FindNumsAppearOnce(data))
# + [markdown] heading_collapsed=true
# # 面试题57:和为s的数字
# # + 题目一:和为s的数字
# 输入一个递增排序的数组和一个数字s,在数组中查找两个数,使得它们的和正好是s。如果有多对数字的和等于s,则输出任意一对即可
# + hidden=true
class Solution:
"""使用两个索引分别指向数组的首尾,指针向中间对撞查找符合要求的数字,对于有多个符合要求的数字对来说,这种方法第一个找到的
必定是两数字只差最大的,因此也是乘积最小的,符合牛客网的附加条件,此外result还具备收集多对符合要求的数字的潜力"""
def FindNumbersWithSum(self, array, tsum):
if not array:
return []
result = []
ahead = len(array) - 1
behind = 0
while ahead > behind:
curSum = array[ahead] + array[behind]
if curSum == tsum:
result.append(array[behind])
result.append(array[ahead])
break
elif curSum > tsum:
ahead -= 1
else:
behind += 1
return result
# test code
# 代码已在牛客网通过测试
# + [markdown] hidden=true
# # + 题目二:和为s的连续正数序列
# 输入一个正数s,打印出所有和为s的连续正数序列(至少含有两个数)。例如,输入15,由于1+2+3+4+5=4+5+6=7+8=15,所以打印出3个连续序列1~5,4~6,7~8。
# + hidden=true
class Solution:
def FindContinuousSequence(self, tsum):
"""和上题相同的思路,指针扩散,设置一个比较小数和较大数,判断其中间的序列和是否符合条件,其中当前序列和的计算参考了
前一序列和,在前一序列和基础上增加big或减少small,这样可以减少计算量"""
result = []
if tsum < 3:
return result
small = 1
big = 2
middle = (1 + tsum) // 2
curSum = small + big
while small < middle:
if curSum == tsum:
result.append([i for i in range(small, big+1)])
while curSum > tsum and small < middle:
curSum -= small
small += 1
if curSum == tsum:
result.append([i for i in range(small, big + 1)])
big += 1
curSum += big
return result
# test code
# 代码已在牛客网通过测试
# + [markdown] heading_collapsed=true
# # 面试题58:翻转字符串
# # + 题目一:翻转单词顺序
# 输入一个英文句子,翻转句子中单词的顺序,但单词内字符的顺序不变。为简单起见,标点符号和普通字母一样处理。例如输入字符串"I am a student",则输出"student a am I"。
# + hidden=true
class Solution:
"""先使用reverse翻转整个句子,再将单词逐个恢复过来"""
def Reverse(self,data, pBegin, pEnd):
if not data:
raise Exception("输入数据为空")
while pBegin < pEnd and pEnd < len(data):
temp = data[pBegin]
data[pBegin] = data[pEnd]
data[pEnd] = temp
pBegin += 1
pEnd -= 1
def ReverseSentence(self, s):
if not s:
return ''
data = list(s)
pBegin = 0
pEnd = 0
self.Reverse(data, pBegin, len(data) - 1)
while pBegin < len(data):
if data[pBegin] == ' ':
pBegin += 1
pEnd += 1
elif pEnd == len(data) or data[pEnd] == ' ':
self.Reverse(data, pBegin, pEnd - 1)
pBegin = pEnd
else:
pEnd += 1
return ''.join(data)
# test code
# 代码已在牛客网通过测试
# + [markdown] hidden=true
# # + 题目二:左旋字符串。
# 字符串的左旋转操作是把字符串的前面若干个字符转移到字符串的尾部。请定义一个函数实现字符串左旋转操作的功能。比如,输入字符串"abcdefg"和数字2,该函数将返回左旋转两位得到的结果"cdefgab"
# + hidden=true
class Solution:
"""和题目一的思路一致,将字符串按n分成两段,先分别翻转,在整体翻转即可达到效果"""
def Reverse(self,data, pBegin, pEnd):
if not data:
raise Exception("输入数据为空")
while pBegin < pEnd and pEnd < len(data):
temp = data[pBegin]
data[pBegin] = data[pEnd]
data[pEnd] = temp
pBegin += 1
pEnd -= 1
def LeftRotateString(self, s, n):
data = s
if s:
length = len(s)
if length > 0 and n > 0 and n < length:
data = list(s)
pFirstStart = 0
pFirstEnd = n - 1
pSecondStart = n
pSecondEnd = length - 1
self.Reverse(data, pFirstStart, pFirstEnd)
self.Reverse(data, pSecondStart, pSecondEnd)
self.Reverse(data, pFirstStart, pSecondEnd)
return ''.join(data)
# test code
# 代码已在牛客网通过测试
# + [markdown] heading_collapsed=true
# # 面试题59:队列的最大值
# # + 题目一:滑动窗口的最大值
# 给定一个数组和滑动窗口的大小,请找出所有滑动窗口里的最大值。例如,如果输入数组{2,3,4,2,6,2,5,1}以及滑动窗口大小3,那么一共存在6个滑动窗口,他们的最大值分别是{4,4,6,6,6,5}。
# + hidden=true
class Solution:
def maxInWindows(self, num, size):
"""使用双端队列,维持队列顶端始终是滑动窗口的最大值的索引位置"""
result = []
if len(num) >= size and size >= 1:
index = []
for i in range(size): # 第一段循环填充满第一个滑动窗口
while index and num[i] >= num[index[-1]]:
index.pop(-1)
index.append(i)
for i in range(size, len(num)): # 第二段循环开始记录每个滑动窗口的最大值
result.append(num[index[0]])
while index and num[i] >= num[index[-1]]:
index.pop(-1)
if index and index[0] <= int(i - size): # 若当前最大值已滑出窗口,则将其删除
index.pop(0)
index.append(i)
result.append(num[index[0]])
return result
# test code
# 代码已在牛客网通过测试
# + [markdown] hidden=true
# # + 题目二:队列的最大值
# 请定义一个队列并实现函数max得到队列里的最大值,要求函数max,push_back和pop_front的时间复杂度都是O(1)。
# + hidden=true
class Solution:
"""思路类似题目一"""
def __init__(self):
self.data = []
self.maximums = []
self.currentIndex = 0
def push_back(self, number):
while self.maximums and number >= self.maximums[-1][0]:
self.maximums.pop(-1)
self.data.append([number, self.currentIndex])
self.maximums.append([number, self.currentIndex])
self.currentIndex += 1
def pop_front(self):
if not self.maximums:
raise Exception('queue is empty')
if self.maximums[0][1] == self.data[0][1]:
self.maximums.pop(0)
self.data.pop(0)
def maxNumber(self):
if not self.maximums:
raise Exception("queue is empty")
return self.maximums[0][0]
# test code
a = Solution()
data = [2,3,4,2,6,2,5,1]
for i in data:
a.push_back(i)
print(a.maxNumber())
if len(a.data) >= 3:
a.pop_front()
# + [markdown] heading_collapsed=true
# # 面试题60:n个骰子的点数
# 题目:把n个骰子仍在地上,所有骰子朝上一面的点数之和为s。输入n,打印出s所有可能的值出现的概率。
# + hidden=true
class Solution:
"""递归的实现算法思路虽然清晰,但算法复杂度高,故而仅实现基于循环的算法,使用两个辅助数组来存储中间结果,多出一个骰子,其增加
的结果的频次是数组前6个数字之和,基于此利用循环来模拟实现"""
def __init__(self):
self.g_maxValue = 6
def probability(self, number):
if number < 1:
return
pProbabilities = [[0]*(self.g_maxValue * number + 1),[0]*(self.g_maxValue * number + 1)]
flag = 0
for i in range(1,self.g_maxValue + 1):
pProbabilities[flag][i] = 1
for j in range(2, number + 1):
for i in range(j):
pProbabilities[1-flag][i] = 0
for i in range(j, self.g_maxValue*j + 1):
pProbabilities[1-flag][i] = 0
k = 1
while k <= i and k <= self.g_maxValue:
pProbabilities[1-flag][i] += pProbabilities[flag][i-k]
k += 1
flag = 1 - flag
total = pow(self.g_maxValue, number)
return [pProbabilities[flag][i]/total for i in range(number, self.g_maxValue*number + 1)]
# test code
a = Solution()
result = a.probability(3)
print(result)
# + [markdown] heading_collapsed=true
# # 面试题61:扑克牌中的顺子
# 题目:从扑克牌中随机抽出5张牌,判断是不是一个顺子,即这5张牌是不是连续的。2~10为数字本身,A为1,J为11,Q为12,K为13,而大、小王可以看成任意数字。
# + hidden=true
class Solution:
def IsContinuous(self, numbers):
"""先将数组进行排序,计算0的个数和间隔的长度,若间隔的长度大于0的个数,则说明使用大小王也无法将五张牌连成顺子,
另外检测到对子则直接判断不是顺子"""
if not numbers:
return False
numbers.sort()
numberOfZero = 0
numberOfGap = 0
for i in numbers:
if i > 0:
break
numberOfZero += 1
small = numberOfZero
big = small + 1
while big < len(numbers):
if numbers[small] == numbers[big]:
return False
numberOfGap += numbers[big] - numbers[small] - 1
small = big
big += 1
return False if numberOfGap > numberOfZero else True
# test code
# 代码已在牛客网通过测试
# + [markdown] heading_collapsed=true
# # 面试题62:圆圈中最后剩下的数字
# 题目:0,1,...,n-1这n个数字排成一个圆圈,从数字0开始,每次从这一个圆圈里删除第m个数字。求出这个圆圈里剩下的最后一个数字。
# + hidden=true
class Solution:
"""此为经典的约瑟夫环问题,可以采用环形链表模拟圆圈进行求解,但此方法时间复杂度为O(mn),空间复杂度O(n)算法效率不高。
另一种方法是找出被删数字的规律,直接计算圆圈中剩下的数字,此方法需要进行数学分析建模,过程较为复杂,但算法效率一般比较高
以下代码根据数学建模后的递归公式写成,算法时间复杂度为O(n),空间复杂度O(1),具体建模过程详见剑指offer,P302-303。"""
def LastRemaining_Solution(self, n, m):
if n < 1 or m < 1:
return -1
last = 0
for i in range(2, n+1):
last = (last + m) % i
return last
# test code
# 代码已在牛客网通过测试
# + [markdown] heading_collapsed=true
# # 面试题63:股票的最大利润
# 题目:假设把某股票的价格按照时间先后顺序存储在数组中,请问买卖该股票一次可能获得的最大利润是多少?例如,一只股票在某些时间节点的价格为{9,11,8,5,7,12,16,14}。如果我们能在价格为5的时候买入,并在价格为16时卖出,则能收获最大的利润11.
# + hidden=true
class Solution:
def MaxDiff(self, numbers):
"""maxdiff存储扫描过的数字中最大利润,minNumber存储扫描过的数字中的最小数字,循环遍历整个数组,
最后得到的必定是最大的差值,即最大利润"""
if len(numbers) < 2:
return 0
minNumber = numbers[0]
maxDiff = numbers[1] - minNumber
for i in range(2, len(numbers)):
if numbers[i-1] < minNumber:
minNumber = numbers[i-1]
currentDiff = numbers[i] - minNumber
if currentDiff > maxDiff:
maxDiff = currentDiff
return maxDiff
# test code
a = Solution()
data = [9,11,8,5,7,12,16,14]
a.MaxDiff(data)
# + [markdown] heading_collapsed=true
# # 面试题64:求1+2+...+n
# 题目:求1+2+...+n,要求不能使用乘除法,for,while,if,else,switch,case等关键字及条件判断语句。
# + hidden=true
class Solution:
"""通过构造函数绕过绕过条件判断,构造出递归终止条件,通过递归实现循环相加。剑指offer中四种方法(构造函数,虚函数,函数指针,
模板类型)都是绕开循环递归相加的思路"""
def __init__(self):
def sum_stop(n):
return 0
self.fun_list = [sum_stop]
def Sum_Solution(self, n):
self.fun_list.append(self.Sum_Solution)
return self.fun_list[bool(n)](n-1) + n
# test code
# 代码已在牛客网通过测试
# + [markdown] heading_collapsed=true
# # 面试题65:不用加减乘除做加法
# 题目:写一个函数,求两整数之和,要求在函数体内不得使用+-*/四则运算符号
# + hidden=true
class Solution:
"""使用异或运算,计算无进位时的结果,使用按位与和左移运算来计算进位数字。由于python在数字即将溢出时会自动使用大数表示,因此
需要对求和结果判断溢出"""
def Add(self, num1, num2):
while num2 != 0:
temp = num1 ^ num2
num2 = (num1 & num2) << 1
num1 = temp & 0xFFFFFFFF
return num1 if num1 >> 31 == 0 else ~(num1 ^ 0xffffffff)
# test code
# 代码已在牛客网通过测试
# + [markdown] heading_collapsed=true
# # 面试题66:构建乘积数组
# 题目:给定一个数组A[0,1,...,n-1],请构建一个数组B[0,1,...,n-1],其中B中的元素$$ B[i]=A[0]\times A[1]\times ...\times A[i-1] \times A[i+1] \times ... \times A[n-1] $$不能使用除法
# + hidden=true
class Solution:
"""将B[i]按照A[i-1]和A[i+1]分成前面乘积C[i]和后面乘积D[i]两部分,对于C[i]=C[i-1]*A[i-1],D[i]=D[i+1]*A[i+1],
因此可以采用从沿着i增加的方向循环计算C[i],再沿着i减少的方向计算D[i],最终获得B"""
def multiply(self, A):
length = len(A)
result = [None]*length
if length <= 1:
return []
# calculate C[i]
result[0] = 1
for i in range(1,length):
result[i] = result[i-1] * A[i-1]
# calculate D[i]*C[i]
temp = 1
for i in range(length-2, -1, -1):
temp *= A[i+1]
result[i] *= temp
return result
# test code
# 代码已在牛客网通过测试
# + [markdown] heading_collapsed=true
# # 面试题67:把字符串转换成整数
# 题目:将一个字符串转换成一个整数(实现Integer.valueOf(string)的功能,但是string不符合数字要求时返回0),要求不能使用字符串转换整数的库函数。 数值为0或者字符串不是一个合法的数值则返回0。
# + hidden=true
class Solution:
def __init__(self):
"""设置输入是否符合要求指示标志"""
self.g_nStatus = True
def StrToInt(self, s):
self.g_nStatus = False
num = 0
if s:
minus = False
i = 0
if s[i] == '+':
i += 1
elif s[i] == '-':
i += 1
minus = True
if i < len(s):
num = self.StrToIntCore(s, i, minus)
return int(num)
def StrToIntCore(self, s, i, minus):
num = 0
while i < len(s):
if s[i] >= '0' and s[i] <= '9':
flag = -1 if minus else 1
num = num * 10 + flag * (ord(s[i]) - ord('0'))
if (not minus and num > 0x7fffffff) or (minus and num < -0x80000000): # 判断是否溢出
num = 0
break
i += 1
else:
num = 0
break
if i == len(s):
self.g_nStatus = True
return num
# test code
#代码已在牛客网通过测试
# -
# # 面试题68:树中两个节点的最低公共祖先
# 题目:输入树的根节点和之中两个节点,求两个节点的最低公共祖先
# + 思路一:判定为二叉搜索树
# 若是二叉搜索树,则通过比较两节点和根节点值的大小,来判断其位于根节点的左子树或右子树,递归判断根节点的子节点,若两节点分别位于子节点(包括根节点)的两侧,则该节点为两节点的最低公共祖先
# + 思路二:若不是二叉搜索树,且有指向父节点的指针
# 若有指向父节点的指针,该问题可转换为求两个链表的第一个公共节点,该链表的尾节点都是指向根节点,可参考两链表的第一个公共节点的解法
# + 思路三:若只是普通树,且没有指向父节点的指针
# 若只是普通树,则可采用深度优先遍历,找到两节点到根节点的路径,将其转换为求两链表的第一个公共节点。以下为此思路的代码(其中测试代码以下图为例):
# 
class TreeNode:
def __init__(self, x):
self.val = x
self.children = []
class Solution:
def GetNodePath(self, pRoot, pNode, path):
"""获取pNode到pRoot的路径,通过修改list类型path返回值"""
if pRoot == pNode:
return True
path.append(pRoot)
found = False
i = 0
while not found and i < len(pRoot.children):
found = self.GetNodePath(pRoot.children[i], pNode, path)
i += 1
if not found:
path.pop(-1)
return found
def GetLastCommonNode(self, path1, path2):
"""遍历两个list模拟的链表,找出最后一个相同的节点"""
i = 0
pLast = None
while i < len(path1) and i < len(path2):
if path1[i] == path2[i]:
pLast = path1[i]
i += 1
return pLast
def GetLastCommonParent(self, pRoot, pNode1, pNode2):
if pRoot == None or pNode1 == None or pNode2 == None:
return None
path1 = []
path2 = []
self.GetNodePath(pRoot, pNode1, path1)
self.GetNodePath(pRoot, pNode2, path2)
return self.GetLastCommonNode(path1, path2)
# test code
pRoot = TreeNode('a')
pRoot.children.append(TreeNode('b'))
pRoot.children.append(TreeNode('c'))
temp = pRoot.children[0]
temp.children.append(TreeNode('d'))
temp.children.append(TreeNode('e'))
temp = temp.children[0]
temp.children.append(TreeNode('f'))
temp.children.append(TreeNode('g'))
temp = pRoot.children[0].children[1]
temp.children.append(TreeNode('h'))
temp.children.append(TreeNode('i'))
temp.children.append(TreeNode('j'))
a = Solution()
reuslt = a.GetLastCommonParent(pRoot, temp.children[0], pRoot.children[0].children[0].children[0])
print(reuslt.val if reuslt != None else reuslt)
| 99,019 |
/examples/scaling-criteo/02-03b-ETL-with-NVTabular-Training-with-PyTorch.ipynb | bb8229beb63faec923dec01d6bdcf52b8131857c | [
"Apache-2.0"
] | permissive | gabrielspmoreira/NVTabular | https://github.com/gabrielspmoreira/NVTabular | 0 | 0 | Apache-2.0 | 2020-11-17T20:17:02 | 2020-11-16T09:30:37 | null | Jupyter Notebook | false | false | .py | 26,866 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# +
# Copyright 2020 NVIDIA Corporation. All Rights Reserved.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#|
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
# ==============================================================================
# -
# # Criteo Example
# Here we'll show how to use NVTabular first as a preprocessing library to prepare the [Criteo Display Advertising Challenge](https://www.kaggle.com/c/criteo-display-ad-challenge) dataset, and then as a dataloader to train a FastAI model on the prepared data. The large memory footprint of the Criteo dataset presents a great opportunity to highlight the advantages of the online fashion in which NVTabular loads and transforms data.
#
# ### Data Prep
# Before we get started, make sure you've run the [optimize_criteo](https://github.com/NVIDIA/NVTabular/blob/main/examples/optimize_criteo.ipynb) notebook, which will convert the tsv data published by Criteo into the parquet format that our accelerated readers prefer. It's fair to mention at this point that that notebook will take around 30 minutes to run. While we're hoping to release accelerated csv readers in the near future, we also believe that inefficiencies in existing data representations like csv are in no small part a consequence of inefficiencies in the existing hardware/software stack. Accelerating these pipelines on new hardware like GPUs may require us to make new choices about the representations we use to store that data, and parquet represents a strong alternative.
# +
import os
from time import time
import re
import glob
import warnings
# tools for data preproc/loading
import torch
import rmm
import nvtabular as nvt
from nvtabular.ops import Normalize, Categorify, LogOp, FillMissing, Clip, get_embedding_sizes
from nvtabular.loader.torch import TorchAsyncItr, DLDataLoader
from nvtabular.utils import device_mem_size, get_rmm_size
# tools for training
from fastai.basics import Learner
from fastai.tabular.model import TabularModel
from fastai.tabular.data import TabularDataLoaders
from fastai.metrics import RocAucBinary, APScoreBinary
from fastai.callback.progress import ProgressCallback
# -
# ### Initializing the Memory Pool
# For applications like the one that follows where RAPIDS will be the only workhorse user of GPU memory and resource, a best practice is to use the RAPIDS Memory Manager library `rmm` to allocate a dedicated pool of GPU memory that allows for fast, asynchronous memory management. Here, we'll dedicate 80% of free GPU memory to this pool to make sure we get the most utilization possible.
rmm.reinitialize(pool_allocator=True, initial_pool_size=get_rmm_size(0.8 * device_mem_size(kind='free')))
# ### Dataset and Dataset Schema
# Once our data is ready, we'll define some high level parameters to describe where our data is and what it "looks like" at a high level.
# +
# define some information about where to get our data
INPUT_DATA_DIR = os.environ.get('INPUT_DATA_DIR', '/raid/criteo/tests/crit_int_pq')
OUTPUT_DATA_DIR = os.environ.get('OUTPUT_DATA_DIR', '/raid/criteo/tests/test_dask') # where we'll save our procesed data to
BATCH_SIZE = int(os.environ.get('BATCH_SIZE', 800000))
PARTS_PER_CHUNK = int(os.environ.get('PARTS_PER_CHUNK', 2))
NUM_TRAIN_DAYS = 23 # number of days worth of data to use for training, the rest will be used for validation
output_train_dir = os.path.join(OUTPUT_DATA_DIR, 'train/')
output_valid_dir = os.path.join(OUTPUT_DATA_DIR, 'valid/')
# ! mkdir -p $output_train_dir
# ! mkdir -p $output_valid_dir
# -
# ! ls $INPUT_DATA_DIR
fname = 'day_{}.parquet'
num_days = len([i for i in os.listdir(INPUT_DATA_DIR) if re.match(fname.format('[0-9]{1,2}'), i) is not None])
train_paths = [os.path.join(INPUT_DATA_DIR, fname.format(day)) for day in range(NUM_TRAIN_DAYS)]
valid_paths = [os.path.join(INPUT_DATA_DIR, fname.format(day)) for day in range(NUM_TRAIN_DAYS, num_days)]
# ### Preprocessing
# At this point, our data still isn't in a form that's ideal for consumption by neural networks. The most pressing issues are missing values and the fact that our categorical variables are still represented by random, discrete identifiers, and need to be transformed into contiguous indices that can be leveraged by a learned embedding. Less pressing, but still important for learning dynamics, are the distributions of our continuous variables, which are distributed across multiple orders of magnitude and are uncentered (i.e. E[x] != 0).
#
# We can fix these issues in a conscise and GPU-accelerated manner with an NVTabular `Workflow`. We define our operation pipelines on `ColumnGroups` (list of column names). Then, we initialize the NVTabular `Workflow` and collect train dataset statistics with `.fit()` and apply the transformation to the train and valid dataset with `.transform()`. NVTabular `ops` can be chained with the overloaded `>>` operator.
#
# #### Frequency Thresholding
# One interesting thing worth pointing out is that we're using _frequency thresholding_ in our `Categorify` op. This handy functionality will map all categories which occur in the dataset with some threshold level of infrequency (which we've set here to be 15 occurrences throughout the dataset) to the _same_ index, keeping the model from overfitting to sparse signals.
# #### Defining the operation pipelines
# 1. Categorical input features (`CATEGORICAL_COLUMNS`) are `Categorify` with frequency treshold of 15
# 2. Continuous input features (`CONTINUOUS_COLUMNS`) filled in missing values, clipped, applied logarithmn and normalized
# +
# define our dataset schema
CONTINUOUS_COLUMNS = ['I' + str(x) for x in range(1,14)]
CATEGORICAL_COLUMNS = ['C' + str(x) for x in range(1,27)]
LABEL_COLUMNS = ["label"]
cat_features = CATEGORICAL_COLUMNS >> Categorify(freq_threshold=15, out_path=OUTPUT_DATA_DIR)
cont_features = CONTINUOUS_COLUMNS >> FillMissing() >> Clip(min_value=0) >> LogOp() >> Normalize()
features = cat_features + cont_features + LABEL_COLUMNS
# -
# We can visualize the pipeline with `graphviz`.
features.graph
# We initialize a NVTabular `Workflow` with our pipelines.
workflow = nvt.Workflow(features)
# Now instantiate dataset's to partition our dataset (which we couldn't fit into GPU memory)
train_dataset = nvt.Dataset(train_paths, part_mem_fraction=0.2)
valid_dataset = nvt.Dataset(valid_paths, part_mem_fraction=0.2)
# Now run them through our workflows to collect statistics on the train set, then transform and save to parquet files.
# %%time
workflow.fit(train_dataset)
# Next, we apply the transformation to the train and valid dataset and persist it to disk.
# %%time
workflow.transform(train_dataset).to_parquet(output_path=output_train_dir,
shuffle=nvt.io.Shuffle.PER_PARTITION,
out_files_per_proc=5)
# %%time
workflow.transform(valid_dataset).to_parquet(output_path=output_valid_dir)
# And just like that, we have training and validation sets ready to feed to a model!
# ## Deep Learning
# ### Data Loading
# We'll start by using the parquet files we just created to feed an NVTabular `TorchAsyncItr`, which will loop through the files in chunks. First, we'll reinitialize our memory pool from earlier to free up some memory so that we can share it with PyTorch.
rmm.reinitialize(pool_allocator=True, initial_pool_size=get_rmm_size(0.3 * device_mem_size(kind='free')))
train_paths = glob.glob(os.path.join(output_train_dir, "*.parquet"))
valid_paths = glob.glob(os.path.join(output_valid_dir, "*.parquet"))
train_data = nvt.Dataset(train_paths, engine="parquet", part_mem_fraction=0.04/PARTS_PER_CHUNK)
valid_data = nvt.Dataset(valid_paths, engine="parquet", part_mem_fraction=0.04/PARTS_PER_CHUNK)
train_data_itrs = TorchAsyncItr(
train_data,
batch_size=BATCH_SIZE,
cats=CATEGORICAL_COLUMNS,
conts=CONTINUOUS_COLUMNS,
labels=LABEL_COLUMNS,
parts_per_chunk=PARTS_PER_CHUNK
)
valid_data_itrs = TorchAsyncItr(
valid_data,
batch_size=BATCH_SIZE,
cats=CATEGORICAL_COLUMNS,
conts=CONTINUOUS_COLUMNS,
labels=LABEL_COLUMNS,
parts_per_chunk=PARTS_PER_CHUNK
)
def gen_col(batch):
return (batch[0], batch[1], batch[2].long())
train_dataloader = DLDataLoader(train_data_itrs, collate_fn=gen_col, batch_size=None, pin_memory=False, num_workers=0)
valid_dataloader = DLDataLoader(valid_data_itrs, collate_fn=gen_col, batch_size=None, pin_memory=False, num_workers=0)
databunch = TabularDataLoaders(train_dataloader, valid_dataloader)
# Now we have data ready to be fed to our model online!
# ### Training
# One extra handy functionality of NVTabular is the ability to use the stats collected by the `Categorify` op to define embedding dictionary sizes (i.e. the number of rows of your embedding table). It even includes a heuristic for computing a good embedding size (i.e. the number of columns of your embedding table) based off of the number of categories.
embeddings = list(get_embedding_sizes(workflow).values())
# We limit the output dimension to 16
embeddings = [[emb[0], min(16, emb[1])] for emb in embeddings]
embeddings
model = TabularModel(emb_szs=embeddings, n_cont=len(CONTINUOUS_COLUMNS), out_sz=2, layers=[512, 256]).cuda()
learn = Learner(databunch, model, loss_func = torch.nn.CrossEntropyLoss(), metrics=[RocAucBinary(), APScoreBinary()])
learning_rate = 1.32e-2
epochs = 1
start = time()
learn.fit(epochs, learning_rate)
t_final = time() - start
total_rows = train_data_itrs.num_rows_processed + valid_data_itrs.num_rows_processed
print(f"run_time: {t_final} - rows: {total_rows} - epochs: {epochs} - dl_thru: {total_rows / t_final}")
| 10,362 |
/data_collection.ipynb | 4fd014a7ad2697d27e72e15424eb79a439e2faa9 | [
"MIT"
] | permissive | fagan2888/dod-ds-overview | https://github.com/fagan2888/dod-ds-overview | 0 | 0 | null | null | null | null | Jupyter Notebook | false | false | .py | 6,595 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# +
from math import sqrt
def canPlace(mat,i,j,n,k):
# check the column and row whether k is present or not
for z in range(0,n):
if mat[i][z]==k or mat[z][j]==k:
return False
# check whether k is present any subgrid of 3x3 or not
# finding the subgrid starting index in which we r check k value
rn=int(sqrt(n))
si=(i//rn)*rn
sj=(j//rn)*rn
for x in range(si,si+rn):
for y in range(sj,sj+rn):
if mat[x][y]==k:
return False
return True
def solveSudoku(mat,i,j,n):
# base case
if i==n:
# Print the matrix
for i in range(n):
for j in range(n):
print(mat[i][j],end=" ")
print("")
return True
# Case Row end
if j==n:
return solveSudoku(mat,i+1,0,n)
# Skip the prefilled cells
if mat[i][j]!=0:
return solveSudoku(mat,i,j+1,n)
# Rec case
# Fill the currrent cell with possible options
for k in range(1,n+1):
if canPlace(mat,i,j,n,k):
mat[i][j]=k
couldWeSolve=solveSudoku(mat,i,j+1,n)
if couldWeSolve:
return True
# Backtracking
mat[i][j]=0
return False
mat=[[0 for i in range(9)] for i in range(9)]
for i in range(9):
z=list(map(int,input().split(' ')))
mat[i]=z
'''
5 3 0 0 7 0 0 0 0
6 0 0 1 9 5 0 0 0
0 9 8 0 0 0 0 6 0
8 0 0 0 6 0 0 0 3
4 0 0 8 0 3 0 0 1
7 0 0 0 2 0 0 0 6
0 6 0 0 0 0 2 8 0
0 0 0 4 1 9 0 0 5
0 0 0 0 8 0 0 7 9
'''
print('')
aa=solveSudoku(mat,0,0,9)
# -
ch += stripped
speech_object = {'url': url,
'title': title,
'date': date,
'speech': speech}
return speech_object
# Now, we can obtain links to each of the respective speeches:
speech_links = []
base = 'https://www.defense.gov/News/Speeches/Customspeechwho/16001/'
for i in range(1,9):
if i == 1:
url = base
else:
url = '{0}?Page={1}'.format(base, i)
response = requests.get(url)
soup = BeautifulSoup(response.text, "html.parser")
links = get_links(soup)
speech_links += links
speech_links = list(set(speech_links))
print('In total, {} speeches were found.'.format(len(speech_links)))
# Now that we have links to the speeches, we can go ahead and obtain the speeches themselves and save them to `.csv`.
speeches = []
for link in list(set(speech_links)):
response = requests.get(link)
soup = BeautifulSoup(response.text, "html.parser")
scraped_speech = process_speech(link, soup)
speeches.append(scraped_speech)
df = pd.DataFrame.from_records(speeches)
# Let's make sure that the speeches are at least 1000 characters in length. Otherwise, it might be junk data.
df = df[df.speech.str.len() > 1000 ]
# Now, we can see that we collected 204 speeches which meet the criteria.
df.shape
# Finally, we can save these speeches as a `.csv` file for future use!
df.to_csv('SecDef_Speeches.csv', index=False)
| 3,277 |
/image_classification.ipynb | 865d657bed4c31640f6dd2252f0f5e3c0d086c19 | [] | no_license | deekshaverma24/Image | https://github.com/deekshaverma24/Image | 0 | 0 | null | null | null | null | Jupyter Notebook | false | false | .py | 23,017 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
#importing dependencies
import numpy as np
import pandas as pd
from matplotlib import pyplot as plt
from sklearn.ensemble import RandomForestClassifier
from sklearn.model_selection import train_test_split
# %matplotlib inline
#using pandas to read the database stored in the same folder
data = pd.read_csv('mnist_train.csv')
# viewing column heads
data.head()
# extracting data from the dataset and viewing them up close
a = data.iloc[3,1:].values
#reshaping the extracted data into a reasonable size
a = a.reshape(28,28).astype('uint8')
plt.imshow(a)
# preparing the data
# seprating labels and data values
df_x = data.iloc[:,1:]
df_y = data.iloc[:,0]
# creating test and train sizes/batches
x_train,x_test,y_train,y_test = train_test_split(df_x,df_y, test_size = 0.2,random_state=4)
#check data
x_train.head()
#check data
y_train.head()
# call rf classifier
rf = RandomForestClassifier(n_estimators=100)
# fit the model
rf.fit(x_train,y_train)
#pediction on test data
pred = rf.predict(x_test)
pred
# +
# check prediction accuracy
s = y_test.values
# calculate number of correctly predicted values
count=0
for i in range (len(pred)):
if pred[i] == s[i]:
count = count+1
# -
count
#total values that to prediction code was run on
len(pred)
# accuracy value
11624/12000
| 1,570 |
/Analysis of IMDB Movie Reviews.ipynb | c830380c2d4f54860803be519459b6878009ac06 | [] | no_license | ShahzebMalikk/Sentiment-Analysis-NLP- | https://github.com/ShahzebMalikk/Sentiment-Analysis-NLP- | 0 | 0 | null | null | null | null | Jupyter Notebook | false | false | .py | 351,866 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Importing libraries
import numpy as np
import pandas as pd
# # Importing Dataset of 20,000 Reviews on IMDB
# +
data = pd.read_excel('Dataset.xlsx')
data.head(10)
# -
# # Checking Dataset Details
data.tail()
data.info()
data.describe()
print(data.shape)
# # sentiment count
data['sentiment'].value_counts()
# # Spliting the training dataset
#train dataset
train_reviews=data.review[:40000]
train_sentiments=data.sentiment[:40000]
#test dataset
test_reviews=data.review[40000:]
test_sentiments=data.sentiment[40000:]
print("train review & train_sentiments" , train_reviews.shape,train_sentiments.shape)
print("test reviews & test_sentiments" , test_reviews.shape,test_sentiments.shape)
# # Text normalization ( Tokenization )
# +
import nltk
from nltk.tokenize.toktok import ToktokTokenizer
from nltk.corpus import stopwords
# +
#nltk.download()
# -
#Tokenization of text
tokenizer=ToktokTokenizer()
#Setting English stopwords
stopword_list=nltk.corpus.stopwords.words('english')
# # Removing all noise text ("cleaning text")
from bs4 import BeautifulSoup
import re
# +
#Removing the html strips
def strip_html(text):
soup = BeautifulSoup(text, "html.parser")
return soup.get_text()
#Removing the square brackets
def remove_between_square_brackets(text):
return re.sub('\[[^]]*\]', '', text)
#Removing the noisy text
def denoise_text(text):
text = strip_html(text)
text = remove_between_square_brackets(text)
return text
#Apply function on review column
data['review']=data['review'].apply(denoise_text)
# -
# # Removing special characters
#Define function for removing special characters
def remove_special_characters(text, remove_digits=True):
pattern=r'[^a-zA-z0-9\s]'
text=re.sub(pattern,'',text)
return text
#Apply function on review column
data['review']=data['review'].apply(remove_special_characters)
# # Text stemming
#Stemming the text
def simple_stemmer(text):
ps=nltk.porter.PorterStemmer()
text= ' '.join([ps.stem(word) for word in text.split()])
return text
#Apply function on review column
data['review']=data['review'].apply(simple_stemmer)
# # Removing stopwords
# +
#set stopwords to english
stop=set(stopwords.words('english'))
print(stop)
#removing the stopwords
def remove_stopwords(text, is_lower_case=False):
tokens = tokenizer.tokenize(text)
tokens = [token.strip() for token in tokens]
if is_lower_case:
filtered_tokens = [token for token in tokens if token not in stopword_list]
else:
filtered_tokens = [token for token in tokens if token.lower() not in stopword_list]
filtered_text = ' '.join(filtered_tokens)
return filtered_text
#Apply function on review column
data['review']=data['review'].apply(remove_stopwords)
# -
# # Normalized train reviews
norm_train_reviews=data.review[:10000]
norm_train_reviews[0]
# # Normalized test reviews
norm_test_reviews=data.review[10000:]
norm_test_reviews[15000]
# # Using only Bags of words model
#
#
# It is used to convert text documents to numerical vectors or bag of words.
from sklearn.feature_extraction.text import CountVectorizer
# +
#Count vectorizer for bag of words
cv=CountVectorizer(min_df=0,max_df=1,binary=False,ngram_range=(1,3))
#transformed train reviews
cv_train_reviews=cv.fit_transform(norm_train_reviews)
#transformed test reviews
cv_test_reviews=cv.transform(norm_test_reviews)
print('BOW_cv_train:',cv_train_reviews.shape)
print('BOW_cv_test:',cv_test_reviews.shape)
#vocab=cv.get_feature_names()-toget feature names
# -
# # Encoding
# Labeling the sentiment text
from sklearn.preprocessing import LabelBinarizer
#labeling the sentient data
lb=LabelBinarizer()
#transformed sentiment data
sentiment_data=lb.fit_transform(data['sentiment'])
print(sentiment_data.shape)
# # Spliting data (Sentiments)
#Spliting the sentiment data
train_sentiments=sentiment_data[:10000]
test_sentiments=sentiment_data[10000:]
print(train_sentiments)
print(test_sentiments)
# # Modelling the dataset using Machine Learning Algorithms
# # 1 : Logistic regression with Bag-of-words
from sklearn.linear_model import LogisticRegression
#training the model
lr=LogisticRegression(penalty='l2',max_iter=200,C=1,random_state=42)
#Fitting the model for Bag of words
lr_bow=lr.fit(cv_train_reviews,train_sentiments)
print(lr_bow)
# ### model performance
lr_bow_predict=lr.predict(cv_test_reviews)
print(lr_bow_predict)
# ### Accuracy of the model
from sklearn.metrics import accuracy_score
#Accuracy score for bag of words
lr_bow_score=accuracy_score(test_sentiments,lr_bow_predict)
print("lr_bow_score :",lr_bow_score)
# ## Classification report
# ### Accuracy, Precision, Recall and F1 score points.
from sklearn.metrics import classification_report,confusion_matrix,accuracy_score
#Classification report for bag of words
lr_bow_report=classification_report(test_sentiments,lr_bow_predict,target_names=['Positive','Negative'])
print(lr_bow_report)
# ## Confusion Matrix
#confusion matrix for bag of words
cm_bow=confusion_matrix(test_sentiments,lr_bow_predict,labels=[1,0])
print(cm_bow)
# # 2 : Building Linear SVM with BOW
from sklearn.linear_model import SGDClassifier
#training the linear svm
svm=SGDClassifier(loss='hinge',max_iter=500,random_state=42)
#fitting the svm for bag of words
svm_bow=svm.fit(cv_train_reviews,train_sentiments)
print(svm_bow)
# ### Model performance on test data
#Predicting the model for bag of words
svm_bow_predict=svm.predict(cv_test_reviews)
print(svm_bow_predict)
# ### Accuracy of the model
#Accuracy score for bag of words
svm_bow_score=accuracy_score(test_sentiments,svm_bow_predict)
print("svm_bow_score :",svm_bow_score)
# ## Classification report
# ### Accuracy, Precision, Recall and F1 score points.
#Classification report for bag of words
svm_bow_report=classification_report(test_sentiments,svm_bow_predict,target_names=['Positive','Negative'])
print(svm_bow_report)
# ### Plotting confusion matrix for it
#confusion matrix for bag of words
cm_bow=confusion_matrix(test_sentiments,svm_bow_predict,labels=[1,0])
print(cm_bow)
# # 3: Multinomial Naive Bayes for bag of words
from sklearn.naive_bayes import MultinomialNB
#training the model
mnb=MultinomialNB()
#fitting the svm for bag of words
mnb_bow=mnb.fit(cv_train_reviews,train_sentiments)
print(mnb_bow)
# ### Model performance on test data
#Predicting the model for bag of words
mnb_bow_predict=mnb.predict(cv_test_reviews)
print(mnb_bow_predict)
# ### Accuracy of the model
#Accuracy score for bag of words
mnb_bow_score=accuracy_score(test_sentiments,mnb_bow_predict)
print("mnb_bow_score :",mnb_bow_score)
# ### classification report
# ### Accuracy, Precision, Recall and F1 score points.
#Classification report for bag of words
mnb_bow_report=classification_report(test_sentiments,mnb_bow_predict,target_names=['Positive','Negative'])
print(mnb_bow_report)
# ### Plot the confusion matrix
#confusion matrix for bag of words
cm_bow=confusion_matrix(test_sentiments,mnb_bow_predict,labels=[1,0])
print(cm_bow)
# # WordNet
import nltk
from nltk.corpus import wordnet #Import wordnet from the NLTK
first_word = wordnet.synset("Travel.v.01")
second_word = wordnet.synset("Walk.v.01")
print('Positive Review Similarity: ' + str(first_word.wup_similarity(second_word)))
first_word = wordnet.synset("Good.n.01")
second_word = wordnet.synset("zebra.n.01")
print('Negative Review Similarity: ' + str(first_word.wup_similarity(second_word)))
# # Extra Work
# # WordCloud for Positive Reviews
from wordcloud import WordCloud,STOPWORDS
import seaborn as sns
import matplotlib.pyplot as plt
#word cloud for positive review words
plt.figure(figsize=(10,10))
positive_text=norm_train_reviews[1]
WC=WordCloud(width=1000,height=500,max_words=500,min_font_size=5)
positive_words=WC.generate(positive_text)
plt.imshow(positive_words,interpolation='bilinear')
plt.show
# # WordCloud for Negative Reviews
#Word cloud for negative review words
plt.figure(figsize=(10,10))
negative_text=norm_train_reviews[8]
WC=WordCloud(width=1000,height=500,max_words=500,min_font_size=5)
negative_words=WC.generate(negative_text)
plt.imshow(negative_words,interpolation='bilinear')
plt.show
# + active=""
# THE END
# -
# # Conclusion
# + active=""
# The accuracy for Logistic Regression is 71% for SVM is 64% and for Naive Bayes is 72% that means the highest we got is 72% using Bag of words vectorizer. we could have used TFIDF for better result but as teacher's requirement we used bag of words. So the Conclusion can be seen in the WORDCLOUD in the positive review wordcloud picture you can see all positive important review words used such as "realism, well, worth, solid, knowlede, fashion, great and many others. Where as for negative the words we got are 'worst, bore, end, mistake, prevent and etc.
#
# Some words have some parts missing it's because we use stemwords and stopwards which removes the part which are not needed for evaluation and other useless words are shown because of somehow not having 100 accuracy.
# -
| 9,343 |
/src/result_plotting.ipynb | 28155204e646ab9ad008ce7ac20a08f7c5de39c7 | [] | no_license | kaan-aytekin/CE690-delay-prediction | https://github.com/kaan-aytekin/CE690-delay-prediction | 0 | 0 | null | null | null | null | Jupyter Notebook | false | false | .py | 2,720,024 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: kaan
# language: python
# name: kaan
# ---
# ## Packages
import os
import pandas as pd
import numpy as np
import seaborn as sns
from matplotlib import pyplot as plt
from matplotlib.pyplot import figure
from warnings import filterwarnings
from pprint import pprint
import pickle
filterwarnings("ignore")
# %matplotlib inline
# ## Global Parameters
ROOT_DIRECTORY = "/home/kaan.aytekin/Thesis"
#ROOT_DIRECTORY = "/Users/kaan.aytekin/Desktop/Kaan/Thesis"
SIMULATION_DURATION = 90 * 60 #Seconds
WARM_UP_PERIOD_DURATION = 15 * 60 #Seconds
COOL_DOWN_PERIOD_DURATION = 15 * 60 #Seconds
DETECTOR_SEPARATION_DISTANCE = 500 #Meter
DETECTOR_COUNT = 21
TOTAL_ROAD_LENGTH = DETECTOR_COUNT * DETECTOR_SEPARATION_DISTANCE
# ### UDFs
# +
def timeit(func):
"""Wrapper function for logging the function duration
Args:
func (function): Function to measure performance
Returns:
function: Function with performance measurement capabilities
"""
from functools import wraps
import datetime as dt
@wraps(func)
def timed(*args, **kwargs):
function_name = func.__name__
start_time = dt.datetime.now()
result = func(*args, **kwargs)
end_time = dt.datetime.now()
execution_duration = (end_time - start_time).total_seconds()
print(
f"Function {function_name!r} executed in {execution_duration:.4f} seconds ({execution_duration/60:.2f} minutes)")
return result
return timed
@timeit
def plot_detector_timelines(df, detector_metric, detector_numbers, simulation_configs):
detector_metric_name_map = {
"flow_vehph": "Flow(Veh/Hr)",
"density_vehpkm": "Density(Veh/Km)",
"avg_speed_kmph": "Speed(Km/Hr)",
"section_travel_time_sec": "Travel Time(Sec)",
"delay_time_sec": "Delay Time(Sec)",
}
detector_metric_name = detector_metric_name_map[detector_metric]
# Data Filtering
simulation_selector = None
for key, value in simulation_configs.items():
if simulation_selector is not None:
simulation_selector = (simulation_selector) & (df[key] == value)
else:
simulation_selector = df[key] == value
detector_selector = None
for detector in detector_numbers:
if detector_selector is not None:
detector_selector = (detector_selector) | (
df["detector_number"] == detector
)
else:
detector_selector = df["detector_number"] == detector
plotting_columns = {
"x": "timestamp",
"y": detector_metric,
"hue": "detector_number",
# "style":"detector_number"
}
plotting_df = df[simulation_selector & detector_selector][
list(set(plotting_columns.values()))
]
# Plotting helpers
max_metric_point = plotting_df[detector_metric].max()
detector_numbers_str = f"{detector_numbers}".strip("[]")
accident_start_time = simulation_configs["accident_start_time"]
accident_duration = simulation_configs["accident_duration"]
accident_detector = int(
np.ceil(simulation_configs["accident_location"] / DETECTOR_SEPARATION_DISTANCE)
)
title = f"{detector_metric_name.split('(')[0]} distribution for Detector(s) {detector_numbers_str}\n Accident Configs: {simulation_configs}\nAccident in detector {accident_detector}"
# Plotting
figure(num=None, figsize=(20, 12), dpi=80, facecolor="w", edgecolor="k")
# ax = sns.lineplot(data=plotting_df,palette=sns.color_palette("Spectral",len(detector_numbers)),**plotting_columns)
ax = sns.lineplot(
data=plotting_df,
palette=sns.color_palette("husl", len(detector_numbers)),
**plotting_columns,
)
ax.set(title=title, xlabel="Timestamp(sec)", ylabel=detector_metric_name)
plt.vlines(
x=accident_start_time,
ymin=0,
ymax=max_metric_point,
colors="blue",
linewidths=3,
)
plt.vlines(
x=accident_start_time + accident_duration,
ymin=0,
ymax=max_metric_point,
colors="blue",
linewidths=3,
)
plt.show()
def prepare_data_for_prediction(df,simulation_configs,scaler,encoder):
df_copy = df.copy()
accident_lane_df = pd.DataFrame(encoder.transform(df_copy[["accident_lane"]]).toarray())
df_copy = df_copy.reset_index(drop=True)
accident_lane_df = accident_lane_df.reset_index(drop=True)
accident_lane_df.columns = encoder.get_feature_names(["accident_lane"])
df_concat = pd.concat([accident_lane_df,df_copy],axis = 1)
simulation_selector = None
for key, value in simulation_configs.items():
if simulation_selector is not None:
simulation_selector = (simulation_selector) & (df_concat[key] == value)
else:
simulation_selector = df_concat[key] == value
df_reduced = df_concat[simulation_selector]
df_model = df_reduced[FEATURE_COLUMNS]
df_model_scaled = scaler.transform(df_model)
return df_model_scaled, df_reduced.target.values
def get_delay_predictions(df,simulation_configs):
df_model, y_real = prepare_data_for_prediction(
df=df,
simulation_configs=simulation_configs,
scaler=min_max_scaler,
encoder=one_hot_encoder
)
y_pred = rf_regressor.predict(df_model)
prediction_error = y_real - y_pred
cumulative_error = np.cumsum(prediction_error)
result_df = pd.DataFrame(
{
"actual_delay": y_real,
"predicted_delay": y_pred,
"prediction_error": prediction_error,
#"cumulative_error": cumulative_error
}
)
return result_df
def plot_delay_predictions(df,simulation_configs):
result_df = get_delay_predictions(df,simulation_configs)
accident_detector = int(
np.ceil(simulation_configs["accident_location"] / DETECTOR_SEPARATION_DISTANCE)
)
detector_number = simulation_configs.get("detector_number")
title = f"Delay prediction for Detector {detector_number}\n Accident Configs: {simulation_configs}\nAccident in detector {accident_detector}"
figure(num=None, figsize=(20, 12), dpi=80, facecolor="w", edgecolor="k")
# ax = sns.lineplot(data=plotting_df,palette=sns.color_palette("Spectral",len(detector_numbers)),**plotting_columns)
ax = sns.lineplot(
data=result_df,
#palette=sns.color_palette("husl", 4),
palette=sns.color_palette("husl", 3),
)
ax.set(title=title, xlabel="Timestamp(sec)", ylabel="Delay Time(sec)")
# -
# ### Load Data
# +
processed_feature_columns_path = os.path.join(
ROOT_DIRECTORY, "data/thesis_data/processed_feature_columns.txt"
)
with open(processed_feature_columns_path, "r") as reader:
FEATURE_COLUMNS = reader.read().split("\n")
top_features_df = pd.read_csv(os.path.join(ROOT_DIRECTORY,"data/thesis_data/top_features.txt"))
selected_features = top_features_df.feature.to_list()
non_feature_columns = [
"simulation_run",
"connected_vehicle_ratio",
"is_accident_simulation",
"accident_lane",
]
df_train = pd.read_csv(
os.path.join(ROOT_DIRECTORY, "data/thesis_data/x_train.csv")
)
y_train = pd.read_csv(
os.path.join(ROOT_DIRECTORY, "data/thesis_data/y_train.csv")
)
df_train["target"] = y_train
df_test = pd.read_csv(
os.path.join(ROOT_DIRECTORY, "data/thesis_data/x_test.csv")
)
y_test = pd.read_csv(
os.path.join(ROOT_DIRECTORY, "data/thesis_data/y_test.csv")
)
df_test["target"] = y_test
df = df_train.append(df_test)
# -
df.head()
# ### Load Model & Scalers
# +
with open(os.path.join(ROOT_DIRECTORY,"model/random_forest_regressor.pkl"),"rb") as file:
rf_regressor = pickle.load(file)
with open(os.path.join(ROOT_DIRECTORY,"model/min_max_scaler.pkl"),"rb") as file:
min_max_scaler = pickle.load(file)
with open(os.path.join(ROOT_DIRECTORY,"model/one_hot_encoder.pkl"),"rb") as file:
one_hot_encoder = pickle.load(file)
# -
# ### Data Exploration (Temp)
def data_slicer(df, simulation_configs):
# Data Filtering
simulation_selector = None
for key, value in simulation_configs.items():
if simulation_selector is not None:
simulation_selector = (simulation_selector) & (df[key] == value)
else:
simulation_selector = df[key] == value
sliced_df = df[simulation_selector]
return sliced_df
simulation_configs = {
"is_accident_simulation" : 1,
#"connected_vehicle_ratio" : 0.6,
"accident_location" : 6636,
"accident_start_time" : 2234,
"accident_duration" : 1492,
"accident_lane" : 1,
"detector_number" : 14
}
data_slicer(df,simulation_configs).head(50)
# ### Plot Prediction Results
# ### Accident vs No Accident
# +
# Accident
simulation_configs = {
"is_accident_simulation" : 1,
"connected_vehicle_ratio" : 0,
"accident_location" : 6932,
"accident_start_time" : 2681,
"accident_duration" : 1245,
"accident_lane" : 1,
"detector_number" : 14
}
plot_delay_predictions(df,simulation_configs)
# -
# No Accident
simulation_configs = {
"simulation_run" : 1,
"is_accident_simulation" : 0,
"connected_vehicle_ratio" : 0,
"accident_location" : 0,
"accident_start_time" : 0,
"accident_duration" : 0,
"accident_lane" : 0,
"detector_number" : 14
}
plot_delay_predictions(df,simulation_configs)
# ### Autonomous Percentage Change
# CV 0%
simulation_configs = {
"is_accident_simulation" : 1,
"connected_vehicle_ratio" : 0,
"accident_location" : 6932,
"accident_start_time" : 2681,
"accident_duration" : 1245,
"accident_lane" : 1,
"detector_number" : 14
}
plot_delay_predictions(df,simulation_configs)
# CV 20%
simulation_configs = {
"is_accident_simulation" : 1,
"connected_vehicle_ratio" : 0.2,
"accident_location" : 6167,
"accident_start_time" : 2588,
"accident_duration" : 849,
"accident_lane" : 1,
"detector_number" : 13
}
plot_delay_predictions(df,simulation_configs)
# CV 40%
simulation_configs = {
"is_accident_simulation" : 1,
"connected_vehicle_ratio" : 0.4,
"accident_location" : 6167,
"accident_start_time" : 2588,
"accident_duration" : 849,
"accident_lane" : 1,
"detector_number" : 13
}
plot_delay_predictions(df,simulation_configs)
# CV 60%
simulation_configs = {
"is_accident_simulation" : 1,
"connected_vehicle_ratio" : 0.6,
"accident_location" : 6167,
"accident_start_time" : 2588,
"accident_duration" : 849,
"accident_lane" : 1,
"detector_number" : 13
}
plot_delay_predictions(df,simulation_configs)
# CV 80%
simulation_configs = {
"is_accident_simulation" : 1,
"connected_vehicle_ratio" : 0.8,
"accident_location" : 6167,
"accident_start_time" : 2588,
"accident_duration" : 849,
"accident_lane" : 1,
"detector_number" : 13
}
plot_delay_predictions(df,simulation_configs)
# ### Incident Lane Change
# Lane 1
simulation_configs = {
"is_accident_simulation" : 1,
"connected_vehicle_ratio" : 0.4,
"accident_location" : 6167,
"accident_start_time" : 2588,
"accident_duration" : 849,
"accident_lane" : 1,
"detector_number" : 13
}
plot_delay_predictions(df,simulation_configs)
# Lane 2
simulation_configs = {
"is_accident_simulation" : 1,
"connected_vehicle_ratio" : 0.4,
"accident_location" : 7825,
"accident_start_time" : 2591,
"accident_duration" : 990,
"accident_lane" : 2,
"detector_number" : 16
}
plot_delay_predictions(df,simulation_configs)
# Lane 3
simulation_configs = {
"is_accident_simulation" : 1,
"connected_vehicle_ratio" : 0.4,
"accident_location" : 7290,
"accident_start_time" : 1076,
"accident_duration" : 674,
"accident_lane" : 3,
"detector_number" : 15
}
plot_delay_predictions(df,simulation_configs)
# ### Accident Duration
# Short
simulation_configs = {
"is_accident_simulation" : 1,
"connected_vehicle_ratio" : 0,
"accident_location" : 6713,
"accident_start_time" : 1119,
"accident_duration" : 607,
"accident_lane" : 1,
"detector_number" : 14
}
plot_delay_predictions(df,simulation_configs)
# Long
simulation_configs = {
"is_accident_simulation" : 1,
"connected_vehicle_ratio" : 0,
"accident_location" : 6636,
"accident_start_time" : 2234,
"accident_duration" : 1492,
"accident_lane" : 1,
"detector_number" : 14
}
plot_delay_predictions(df,simulation_configs)
# ### Possible Configuration Means
df.columns[:20]
df.groupby(by=["is_accident_simulation", "accident_lane", "connected_vehicle_ratio"])["flow_vehph","density_vehpkm","avg_speed_kmph","section_travel_time_sec","delay_time_sec"].mean()
# ### Autonomous Connected Vehicle Incident Local Area Improvement
simulation_configs = {
"is_accident_simulation" : 1,
#"connected_vehicle_ratio" : 0.6,
"accident_location" : 6636,
"accident_start_time" : 2234,
"accident_duration" : 1492,
"accident_lane" : 1,
#"detector_number" : 14
}
local_incident_data = data_slicer(df,simulation_configs)
local_incident_data[
(local_incident_data.detector_number == 13)
| (local_incident_data.detector_number == 14)
| (local_incident_data.detector_number == 15)
].groupby(["connected_vehicle_ratio","detector_number"])["flow_vehph","density_vehpkm","avg_speed_kmph","section_travel_time_sec","delay_time_sec"].mean()
# ### Autonomous Connected Vehicle Incident Global Area Improvement
simulation_configs = {
"is_accident_simulation" : 1,
#"connected_vehicle_ratio" : 0.6,
"accident_location" : 6636,
"accident_start_time" : 2234,
"accident_duration" : 1492,
"accident_lane" : 1,
#"detector_number" : 14
}
local_incident_data = data_slicer(df,simulation_configs)
local_incident_data.groupby(["connected_vehicle_ratio"])["flow_vehph","density_vehpkm","avg_speed_kmph","section_travel_time_sec","delay_time_sec"].sum().to_csv()
simulation_configs = {
"is_accident_simulation" : 1,
"connected_vehicle_ratio": 0,
"accident_location" : 6932,
"accident_start_time" : 2681,
"accident_duration" : 1245,
"accident_lane" : 1,
"detector_number" : 15
}
plot_delay_predictions(df,simulation_configs)
simulation_configs = {
"is_accident_simulation" : 1,
"connected_vehicle_ratio": 0.5,
"accident_location" : 6167,
"accident_start_time" : 2588,
"accident_duration" : 849,
"accident_lane" : 1,
"detector_number" : 15
}
plot_delay_predictions(df,simulation_configs)
df[
(df.connected_vehicle_ratio == 0.5)
][
[
"is_accident_simulation",
"accident_location",
"accident_start_time",
"accident_duration",
"accident_lane"]
].drop_duplicates()
| 14,905 |
/capital_bikeshare_fun.ipynb | 3fea62d28da8a42cc184d10bb38eb25f6d9c0209 | [] | no_license | Aycrazy/capital_bikeshare_optimizer | https://github.com/Aycrazy/capital_bikeshare_optimizer | 0 | 0 | null | null | null | null | Jupyter Notebook | false | false | .py | 897,524 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Python 2
# language: python
# name: python2
# ---
# +
'''
from lxml import html
import requests
def get_category_links(section_url):
html = urlopen(section_url).read()
soup = BeautifulSoup(html, "lxml")
boccat = soup.find("dl", "boccat")
category_links = [BASE_URL + dd.a["href"] for dd in boccat.findAll("dd")]
return category_links
'''
"""index = 'https://en.wikipedia.org/wiki/Watchmen' #'http://www.snackdata.com'
soup = BeautifulSoup(requests.get(index).content, 'html.parser')
#items = soup.find(id='indexholder').find_all('li')
soup.find(id='Story')
#[index + item.find('a')['href'] for item in items]
"""
#import requests
#BeautifulSoup(requests.get(links[0]).content, 'html.parser')
"""
section_url = 'https://en.wikipedia.org/wiki/Watchmen'
html = urlopen(section_url).read()
soup = BeautifulSoup(html, 'lxml')
watch = ''.join(str(soup.findAll('p')[17:22]))
section_url = 'https://en.wikipedia.org/wiki/Saga_(comic_book)'
html = urlopen(section_url).read()
soup = BeautifulSoup(html, 'lxml')
saga = ''.join(str(soup.findAll('p')[16:20]))
section_url = 'https://en.wikipedia.org/wiki/V_for_Vendetta'
html = urlopen(section_url).read()
soup = BeautifulSoup(html, 'lxml')
v = ''.join(str(soup.findAll('p')[15:22]))
"""
# +
# Scraping
from bs4 import BeautifulSoup
from urllib2 import urlopen
def get_text(link_list):
links = []
text_list = list()
for link in link_list:
try:
html = urlopen(link).read()
soup = BeautifulSoup(html, 'lxml')
text_list.append(''.join(str(soup.findAll('p'))))
links.append(link)
except:
print link
return (links, text_list)
links = ['https://en.wikipedia.org/wiki/100_Bullets',
'https://en.wikipedia.org/wiki/2000_AD_(comics)',
'https://en.wikipedia.org/wiki/300_(comics)',
'https://en.wikipedia.org/wiki/A_Contract_with_God',
'https://en.wikipedia.org/wiki/Akira_(manga)',
'https://en.wikipedia.org/wiki/All-Star_Superman',
'https://en.wikipedia.org/wiki/Annihilation_(comics)',
'https://en.wikipedia.org/wiki/Arkham_Asylum:_A_Serious_House_on_Serious_Earth',
'https://en.wikipedia.org/wiki/Astonishing_X-Men',
'https://en.wikipedia.org/wiki/Aya_of_Yop_City',
'https://en.wikipedia.org/wiki/Barefoot_Gen',
'https://en.wikipedia.org/wiki/Batman:_The_Killing_Joke',
'https://en.wikipedia.org/wiki/Batman:_The_Long_Halloween',
'https://en.wikipedia.org/wiki/Batman:_Year_One',
'https://en.wikipedia.org/wiki/Black_Hole_(comics)',
'https://en.wikipedia.org/wiki/Blankets_(comics)',
'https://en.wikipedia.org/wiki/Bone_(comics)',
'https://en.wikipedia.org/wiki/Born_Again_(comics)',
'https://en.wikipedia.org/wiki/Chew_(comics)',
'https://en.wikipedia.org/wiki/Civil_War_(comics)',
'https://en.wikipedia.org/wiki/DMZ_(comics)',
'https://en.wikipedia.org/wiki/Ex_Machina_(comics)',
'https://en.wikipedia.org/wiki/Fables_(comics)',
'https://en.wikipedia.org/wiki/From_Hell',
'https://en.wikipedia.org/wiki/Fun_Home',
'https://en.wikipedia.org/wiki/Ghost_World',
'https://en.wikipedia.org/wiki/Girl_Genius',
'https://en.wikipedia.org/wiki/Hellblazer',
'https://en.wikipedia.org/wiki/Hellboy:_Seed_of_Destruction',
'https://en.wikipedia.org/wiki/Kingdom_Come_(comics)',
'https://en.wikipedia.org/wiki/Krazy_Kat',
'https://en.wikipedia.org/wiki/List_of_Criminal_story_arcs',
'https://en.wikipedia.org/wiki/List_of_Preacher_story_arcs',
'https://en.wikipedia.org/wiki/Locke_%26_Key',
'https://en.wikipedia.org/wiki/Lone_Wolf_and_Cub',
'https://en.wikipedia.org/wiki/Louis_Riel_(comics)',
'https://en.wikipedia.org/wiki/MIND_MGMT',
'https://en.wikipedia.org/wiki/Marvels',
'https://en.wikipedia.org/wiki/Maus',
'https://en.wikipedia.org/wiki/Palestine_(comics)',
'https://en.wikipedia.org/wiki/Persepolis_(comics)',
'https://en.wikipedia.org/wiki/Powers_(comics)',
'https://en.wikipedia.org/wiki/Saga_(comic_book)',
'https://en.wikipedia.org/wiki/Scalped',
'https://en.wikipedia.org/wiki/Scott_Pilgrim',
'https://en.wikipedia.org/wiki/Sin_City',
'https://en.wikipedia.org/wiki/Superman:_Red_Son',
'https://en.wikipedia.org/wiki/Swamp_Thing',
'https://en.wikipedia.org/wiki/The_Authority',
'https://en.wikipedia.org/wiki/The_Dark_Knight_Returns',
'https://en.wikipedia.org/wiki/The_Dark_Phoenix_Saga',
'https://en.wikipedia.org/wiki/The_Galactus_Trilogy',
'https://en.wikipedia.org/wiki/The_Invisibles',
'https://en.wikipedia.org/wiki/The_League_of_Extraordinary_Gentlemen_(comics)',
'https://en.wikipedia.org/wiki/The_Maxx',
'https://en.wikipedia.org/wiki/The_New_Avengers_(comics)',
'https://en.wikipedia.org/wiki/The_Night_Gwen_Stacy_Died',
'https://en.wikipedia.org/wiki/The_Sandman_(Vertigo)',
'https://en.wikipedia.org/wiki/The_Ultimates_(comic_book)',
'https://en.wikipedia.org/wiki/The_Walking_Dead_(comic_book)',
'https://en.wikipedia.org/wiki/Time_(xkcd)',
'https://en.wikipedia.org/wiki/Transmetropolitan',
'https://en.wikipedia.org/wiki/Uncanny_X-Men',
'https://en.wikipedia.org/wiki/V_for_Vendetta',
'https://en.wikipedia.org/wiki/Wanted_(comics)',
'https://en.wikipedia.org/wiki/Watchmen',
'https://en.wikipedia.org/wiki/Y:_The_Last_Man',
]
links, comic_text = get_text(links)
# -
# +
# TFIDF
from nltk.corpus import stopwords
from nltk.tokenize import word_tokenize
from nltk.stem.snowball import SnowballStemmer
from string import punctuation
from sklearn.metrics.pairwise import cosine_similarity
from sklearn.feature_extraction.text import TfidfVectorizer
# +
def clean_text(list_o_text):
docs = [''.join([char if char not in punctuation else ' ' for char in
comic]) for comic in list_o_text]
# remove punctuation from string
docs = [word_tokenize(comic) for comic in docs]
# make string into list of words
# 3. Strip out stop words from each tokenized document.
stop = set(stopwords.words('english'))
stop.update(punctuation)
other_words = ['cite', 'cite_note', 'cite_ref', 'class', 'href', 'id',
'redirect', 'ref', 'refer', 'span', 'sup', 'title', 'wiki']
stop.update(other_words)
docs = [[word for word in words if word.strip(punctuation) not in stop]
for words in docs]
# remove stop words
# Stemming / Lemmatization
# 1. Stem using both stemmers and the lemmatizer
#porter = PorterStemmer()
snowball = SnowballStemmer('english')
#wordnet = WordNetLemmatizer()
#docs_porter = [[porter.stem(word) for word in words] for words in docs]
docs_snowball = [[snowball.stem(word) for word in words] for words in docs]
#docs_wordnet = [[wordnet.lemmatize(word) for word in words] for words in docs]
docs = [' '.join(doc) for doc in docs_snowball]
# for each document, it becomes a long string
return docs
docs = clean_text(comic_text)
# -
tfidf_vectorizer = TfidfVectorizer(stop_words='english')
tfidfed_matrix = tfidf_vectorizer.fit_transform(docs)
# docs must be list of strings
tfidf_vectorizer.vocabulary_
# +
cosine_similarities = cosine_similarity(tfidfed_matrix.todense(),
tfidfed_matrix.todense())
for i, link in enumerate(links):
for j, link in enumerate(links):
print i, j, cosine_similarities[i, j]
# -
print cosine_similarities.shape
print len(tfidf_vectorizer.vocabulary_)
# +
# save the data
import cPickle as pickle
with open('links.pkl', 'w') as f:
pickle.dump(links, f)
with open('vectorizer.pkl', 'w') as f:
pickle.dump(tfidf_vectorizer, f)
with open('matrix.pkl', 'w') as f:
pickle.dump(tfidfed_matrix, f)
with open('comic_text.pkl', 'w') as f:
pickle.dump(comic_text, f)
# -
# +
# load the data
#import glob
#print glob.glob("*.pkl")
import cPickle as pickle
with open('links.pkl') as f:
links = pickle.load(f)
with open('vectorizer.pkl') as f:
tfidf_vectorizer = pickle.load(f)
with open('matrix.pkl') as f:
tfidfed_matrix = pickle.load(f)
# +
# import same as above
from nltk.corpus import stopwords
from nltk.tokenize import word_tokenize
from nltk.stem.snowball import SnowballStemmer
from string import punctuation
from sklearn.metrics.pairwise import cosine_similarity
from sklearn.feature_extraction.text import TfidfVectorizer
# -
# +
# tokenize input string > outputs a list of 1 string
# import stuff from above
def string_cleaner(input_string):
stop = set(stopwords.words('english'))
stop.update(punctuation)
user_input = word_tokenize(input_string)
snowball = SnowballStemmer('english')
user_snowball = [snowball.stem(word) for word in user_input if word
not in stop]
# remove useless words
#lowercase words, keeps only root of words
user = [str(' '.join(user_snowball))]
# converts list of words into string
return user
# -
# +
# recommendation
import numpy as np
def cos_sim_recommender(input_string, tfidfed_matrix, links):
user = string_cleaner(input_string)
recommend = cosine_similarity(tfidf_vectorizer.transform(user).todense(),
tfidfed_matrix.todense())
# x-axis is the original data, y-axis is the query (raw_input) you put in
# docs must be list of strings
title_index = np.argmax(recommend)
# find max similarity
return links[title_index].split('/')[-1]
# recommendation!
"""
new_docs = ['Saga has two planets where poorer and richer fighter with smelly babies',
'watchmen has heroes with silky spectres and masked man',
'vendettta vengeance bombing evey in London',
"Four months later, V breaks into Jordan Tower, the home of Norsefire's propaganda department"]
"""
cos_sim_recommender(raw_input('type what you want> '), tfidfed_matrix, links)
# -
# +
# 1. Apply k-means clustering to the articles.pkl
from sklearn.cluster import KMeans
def make_kclusters(tfidf_vectorizer, n_clusters = 8):
#X is matrix
# features is list of words
kmeans = KMeans(n_clusters=n_clusters)
kmeans.fit(tfidfed_matrix)
features = tfidf_vectorizer.get_feature_names()
# 2. Print out the centroids.
#print "cluster centers:"
#print kmeans.cluster_centers_
# 3. Find the top 10 features for each cluster.
top_centroids = kmeans.cluster_centers_.argsort()[:,-1:-19:-1]
print "top features for each cluster:"
for num, centroid in enumerate(top_centroids):
print "%d: %s" % (num, ", ".join(features[i] for i in centroid))
return kmeans
kmeans = make_kclusters(tfidf_vectorizer)
# -
# Print KMean Clusters
import numpy as np
def print_kclusters(kmeans = kmeans):
titles = np.array([link.split('/')[-1] for link in links])
for index_num, label in enumerate(set(kmeans.labels_)): #index_num isn't true label
indices = np.where(kmeans.labels_ == label)[0]
print index_num
for index in indices:
print titles[index]
print ""
print_kclusters()
# +
# Hierarchical clustering
from scipy.spatial.distance import pdist, squareform
from scipy.cluster.hierarchy import linkage, dendrogram
import matplotlib.pyplot as plt
def draw_dendrogram(tfidf_matrix):
# distxy = squareform(pdist(tfidf_matrix.todense(),
# metric='cosine'))
link = linkage(tfidf_matrix.todense(), method='complete', metric='cosine')
dendro = dendrogram(link, color_threshold=1.5, leaf_font_size=9, labels=
[link.split('/')[-1] for link in links], leaf_rotation=90)
plt.show()
draw_dendrogram(tfidfed_matrix)
# -
# +
# Get from 1 known comic book to a recommendation
#print distxy[9, 0] # numbers Bone > Saga
#print distxy[9, 4] # dendro Bone > Time
#print distxy[9,:]
#print distxy[0, 2] # distxy: Saga > Watchmen
#print distxy[0, 22] # dendro: Saga > Maus
#print distxy[0]
# comic to comic
def cos_sim_c2c(input_string, rejected_comics=[], how_many = 3):
titles = np.array([link.split('/')[-1] for link in links])
try:
which_comic = np.where(titles == input_string)[0][0]
except:
return 'Your preferred comic title is not in this database'
distxy = squareform(pdist(tfidfed_matrix.todense(), metric='cosine'))
closest_comics = titles[np.argsort(distxy[which_comic])][1:]
best_n_comics = []
for comic in closest_comics:
if comic in rejected_comics:
continue
else:
best_n_comics.append(comic)
if len(best_n_comics) == how_many:
return best_n_comics
return best_n_comics
good_comic = raw_input('What comic do you want a similar one? ')
bad_comics = raw_input('Comics you hate? Separate by commas ').split(',')
bad_comics = [comic.strip() for comic in bad_comics]
cos_sim_c2c(good_comic, bad_comics)
# +
# Maus > Saga_(comic_book), Watchmen, The_Sandman_(Vertigo)
#distxy = squareform(pdist(tfidfed_matrix.todense(), metric='cosine'))
#np.array([link.split('/')[-1] for link in links])[
# np.argsort(distxy[22])]
# +
import random
# cosine similarity random comic to comic
def cos_sim_rc2c():
titles = np.array([link.split('/')[-1] for link in links])
random_comic = random.choice(titles)
over_recommended_comics = 'The_Sandman_(Vertigo), Watchmen, Saga_(comic_book)'
output = "A random comic: " + random_comic + "; Similar comics: "
return output + ', '.join(cos_sim_c2c(random_comic, '', how_many = 3))
# -
cos_sim_rc2c()
# +
# constructing NMF
from sklearn.decomposition import NMF
nmf = NMF(n_components=10)
W_sklearn = nmf.fit_transform(tfidfed_matrix)
H_sklearn = nmf.components_
# +
def reconst_mse(target, left, right):
return (np.array(target - left.dot(right))**2).mean()
def describe_nmf_results(document_term_mat = tfidfed_matrix, W = W_sklearn,
H = H_sklearn, n_top_words = 15, vectorizer = tfidf_vectorizer):
feature_words = vectorizer.get_feature_names()
print("Reconstruction error: %f") %(reconst_mse(document_term_mat, W, H))
for topic_num, topic in enumerate(H):
print("Topic %d:" % topic_num)
print(" ".join([feature_words[i] \
for i in topic.argsort()[:-n_top_words - 1:-1]]))
return
describe_nmf_results()
# +
# print W_sklearn.shape, H_sklearn.shape, tfidfed_matrix.shape
#np.where(titles == 'The_Galactus_Trilogy')
# -
def print_nmf_clusters(W_sklearn = W_sklearn, H_sklearn = H_sklearn):
for cluster_index in range(len(H_sklearn)):
titles = np.array([link.split('/')[-1] for link in links])
comics_in_cluster = []
print cluster_index
for ith, comic in enumerate(W_sklearn):
if cluster_index == np.argmax(comic):
print titles[ith]
print ""
print_nmf_clusters()
def word_to_index_in_vectorizer(lst_o_words):
word_indices = []
for word in lst_o_words[0].split():
try:
word_indices.append(tfidf_vectorizer.vocabulary_[word])
except:
continue
return word_indices
# +
# input string > comic book recommendations; like cosine similarity
def nmf_recommender_1(input_string, tfidfed_matrix, links):
user = string_cleaner(input_string)
# get tokenized words from input string
word_indices = word_to_index_in_vectorizer(user)
# for each word, get the index from vectorizer
average_topics = [0] * H_sklearn.shape[0]
for index in range(len(average_topics)):
average_topics[index] = H_sklearn[index][word_indices].mean()
# for each word, get the "average" that the word would appear in
guess = np.argmax(cosine_similarity(average_topics, W_sklearn))
return np.array([link.split('/')[-1] for link in links])[guess]
nmf_recommender_1(raw_input('type what you want> '), tfidfed_matrix, links)
# +
#print W_sklearn.shape
#print np.array(average_topics).reshape((1, -1)).shape
#print np.argmax(cosine_similarity(np.array(average_topics).reshape((1, -1)),
# W_sklearn))
#print tfidf_vectorizer.transform(user).shape
#print tfidfed_matrix.shape
#print cosine_similarity(tfidf_vectorizer.transform(user).todense(),
# tfidfed_matrix.todense()).shape
#W_sklearn[:, index].copy().argsort()[::-1].argsort()
#np.argmin(np.mean(np.array(ranked_comics), axis=0))
#from collections import Counter
#Counter(top_topics)
#np.linalg.lstsq(H_sklearn.T, tfidf_vectorizer.transform(user).todense().reshape((-1, 1)))[0]
# -
def nmf_recommender_2(input_string, tfidfed_matrix=tfidfed_matrix, links=links):
user = string_cleaner(input_string)
# get tokenized words from input string
word_indices = word_to_index_in_vectorizer(user)
# for each word, get the index from vectorizer
top_topics = []
for index in word_indices:
top_topics.append(np.argmax(H_sklearn[:, index]))
# get top topics for each word
ranked_comics = []
for index in top_topics:
order = W_sklearn[:, index].copy().argsort()[::-1]
ranks = order.argsort()
ranked_comics.append(list(ranks))
# rank all comic books for each topic; lower is better
guess = np.argmin(np.mean(np.array(ranked_comics), axis=0))
# find the comic that is relatively low (closest) on each topic
return np.array([link.split('/')[-1] for link in links])[guess]
nmf_recommender_2(raw_input('type in for nmf recommendation> '))
"""
def get_similar_comics_nmf(W_sklearn, which_comic):
order_topics_for_comics = get_sorted_topics_matrix(W_sklearn)
this_comic_topics_index = list(W_sklearn[which_comic].copy().argsort()[::-1])
# sorted importance for this comic
possible_options = [np.array(range(len(order_topics_for_comics)))]
# generates which comics are similar by FP growth
for i, topic_index in enumerate(this_comic_topics_index):
where = np.where(order_topics_for_comics[possible_options[-1]][:, i] == topic_index)
possible_options.append(possible_options[-1][where[0]])
if len(possible_options) == 2:
return list(possible_options[-1])
elif len(possible_options) == 1:
return list(possible_options[-2])
"""
# creates array where it shows each row is each comics sorted topics in descreasing importance
def get_sorted_topics_matrix(W_sklearn):
order_topics_for_comics = []
for comic in range(len(W_sklearn)):
topics_for_comic = list(W_sklearn[comic].copy().argsort()[::-1])
order_topics_for_comics.append(topics_for_comic)
return np.array(order_topics_for_comics)
def get_similar_comics_nmf(W_sklearn, which_comic):
order_topics_for_comics = get_sorted_topics_matrix(W_sklearn)
this_comic_topics_index = list(W_sklearn[which_comic].copy().argsort()[::-1])
# sorted importance for this comic
possible_options = [np.array(range(len(order_topics_for_comics)))]
# generates which comics are similar by FP growth
for i, topic_index in enumerate(this_comic_topics_index):
where = np.where(order_topics_for_comics[possible_options[-1]][:, i] == topic_index)
possible_options.append(possible_options[-1][where[0]])
if len(possible_options) == 2:
return list(possible_options[-1])
elif len(possible_options) == 1:
return list(possible_options[-2])
# inside cluster nmf comic to comic recommender
def nmf_c2c_in(input_string, rejected_comics = [], how_many = 3):
titles = np.array([link.split('/')[-1] for link in links])
try:
which_comic = np.where(titles == input_string)[0][0]
except:
return 'Your preferred comic title is not in this database'
recommendations = [x for x in get_similar_comics_nmf(W_sklearn, which_comic) if x != which_comic]
#recommendations = get_comic_index(W_sklearn, which_comic, float('inf'))
comic_recommendations = titles[np.array(recommendations)]
np.random.shuffle(comic_recommendations)
print comic_recommendations
best_n_comics = []
for comic in comic_recommendations:
if comic in rejected_comics:
continue
else:
best_n_comics.append(comic)
if len(best_n_comics) == how_many:
return best_n_comics
return best_n_comics
# +
good_comic = raw_input('What comic do you want a similar one? ')
bad_comics = raw_input('Comics you hate? Separate by commas ').split(',')
bad_comics = [comic.strip() for comic in bad_comics]
nmf_c2c_in(good_comic, bad_comics)
# input Scalped
# possible recommendations: ['The_Invisibles' 'Girl_Genius' 'From_Hell'
# 'List_of_Y:_The_Last_Man_story_arcs' 'Hellboy:_Seed_of_Destruction'
# 'Wanted_(comics)' 'V_for_Vendetta' 'List_of_Preacher_story_arcs'
# 'List_of_Criminal_story_arcs'
# 'The_League_of_Extraordinary_Gentlemen_(comics)']
# output
# restrict: List_of_Y:_The_Last_Man_story_arcs, From_Hell, Girl_Genius, List_of_Criminal_story_arcs,
# Hellboy:_Seed_of_Destruction, The_League_of_Extraordinary_Gentlemen_(comics), Wanted_(comics)
# get: V, Preacher, Invisibles
# -
| 21,694 |
/course 1 - Algorithmic toolbox/week5_dynamic_programming1/2_primitive_calculator/primitive calculator.ipynb | 8cb5c866e99fef29725f41f811e02c1bf92b0859 | [] | no_license | prachivishnoi27/Data-Structures-and-Algorithms-specialization-University-of-California-San-Diego | https://github.com/prachivishnoi27/Data-Structures-and-Algorithms-specialization-University-of-California-San-Diego | 1 | 0 | null | 2020-07-11T12:38:00 | 2020-07-07T06:21:50 | null | Jupyter Notebook | false | false | .py | 4,526 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# Определите количество мужчин и женщин среди испытуемых. Обратите внимание, что способ кодирования переменной gender мы не знаем. Воспользуемся медицинским фактом, а именно: мужчины в среднем выше женщин.
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
sns.set();
df = pd.read_csv('../input/cardiovascular-disease-dataset/cardio_train.csv', sep=';')
df.head()
new_df = pd.DataFrame()
new_df['mean_height'] = df.groupby('gender')['height'].mean()
new_df['count'] = df['gender'].value_counts()
new_df
# Верно ли, что мужчины более склонны к употреблению алкоголя, чем женщины?
pd.crosstab(df['gender'], df['alco'])
# Каково различие между процентами курящих мужчин и женщин?
pd.crosstab(df['gender'], df['smoke'], normalize=True)
df[df['gender'] == 1]['smoke'].sum() / df[df['gender'] == 1].shape[0]
df[df['gender'] == 2]['smoke'].sum() / df[df['gender'] == 1].shape[0]
# Какова разница между средними значениями возраста для курящих и некурящих?
df['age'] = (df['age'] / 365).round()
df.groupby('smoke')['age'].mean()
# Создайте новый признак --- BMI (body mass index, индекс массы тела). Для этого разделите вес в килограммах на квадрат роста в метрах. Считается, что нормальные значения ИМТ составляют от 18.5 до 25. Выберите верные утверждения:
#
# (a) Средний ИМТ находится в диапазоне нормальных значений ИМТ.
#
# (b) ИМТ для женщин в среднем выше, чем для мужчин.
#
# (c) У здоровых людей в среднем более высокий ИМТ, чем у людей с ССЗ.
#
# (d) Для здоровых непьющих мужчин ИМТ ближе к норме, чем для здоровых непьющих женщин
df['BMI'] = df['weight'] / ((df['height']/100)**2)
df['BMI'].mean()
df.groupby('gender')['BMI'].mean()
df.groupby('cardio')['BMI'].mean()
df.pivot_table(values=['BMI'], index=['cardio', 'alco', 'gender'], aggfunc='mean')
# Удалите пациентов, у которых диастолическое давление выше систолического. Какой процент от общего количества пациентов они составляли?
df[df['ap_hi'] < df['ap_lo']].shape[0] / df.shape[0]
df.drop(df[df['ap_hi'] < df['ap_lo']].index)
# На сайте Европейского общества кардиологов представлена шкала SCORE. Она используется для расчёта риска смерти от сердечно-сосудистых заболеваний в ближайшие 10 лет.
#
# Рассмотрим верхний правый прямоугольник, который показывает подмножество курящих мужчин в возрасте от 60 до 65 лет (значения по вертикальной оси на рисунке представляют верхнюю границу).
#
# Мы видим значение 9 в левом нижнем углу прямоугольника и 47 в правом верхнем углу. Это означает, что для людей этой возрастной группы с систолическим давлением менее 120 и низким уровнем холестерина риск сердечно-сосудистых заболеваний оценивается примерно в 5 раз ниже, чем для людей с давлением в интервале [160, 180] и высоким уровнем холестерина.
#
# Вычислите аналогичное соотношение для наших данных.
platform_genre_sales = df[(df['ap_hi'] > 120) & (df['ap_hi'] < 180) & (df['age'] > 60) & (df['age'] < 65) & (df['gender'] == 2)].pivot_table(
index='ap_hi',
columns='cholesterol',
values='cardio',
aggfunc=sum).fillna(0).applymap(float)
f, ax = plt.subplots(figsize=(10, 17))
sns.heatmap(platform_genre_sales, annot=True, linewidths=.5, fmt=".1f", ax=ax, yticklabels=5)
# Визуализируйте распределение уровня холестерина для различных возрастных категорий.
plt.figure(figsize=(25, 15))
sns.countplot(y='age', hue='cholesterol', data=df);
# Как распределена переменная BMI? Есть ли выбросы
sns.boxplot(df['BMI']);
# Как соотносятся ИМТ и наличие ССЗ? Придумайте подходящую визуализацию.
df.groupby(['cardio', 'gender'])['BMI'].mean().plot(kind='bar')
plt.ylabel('BMI')
plt.show();
s():
logger.debug(f'Processing {field}')
data_utils.process_field(df_filt, field, hard_rejects, **params)
# Filter out hard rejects
for _, ids in hard_rejects.items():
if len(ids) > 0:
df_filt = df_filt.loc[~df_filt.index.isin(ids)]
logger.info(f'Length of records after hard rejects {len(df_filt):,}')
return df_filt, hard_rejects
def process_soft_rejects(df_filt):
'''
Here we define any logic based on columns that can be null, and for which a validation error would mean we set the column to null
After processing, the data in the original column names will have clean data in them, and columns like '[col_name]_orig' will have the
original value if it failed validation. See data_utils.process_field for a detailed explanation
Parameters:
df_filt (DataFrame): our data after processing hard rejects (modified in place)
Returns:
soft_rejects (dict of column name, indices): column names and row indices of anything that failed some validation and
had to be set to null because of it
'''
soft_rejects = defaultdict(set)
# Generic cosmetic changes (post-processing)
capitalize_first = lambda series: series.str.title()
upper_case = lambda series: series.str.upper()
# Block we want to extract the hidden house number from the street location so we can analyze crime by street
def block_num_addr(df, blocks):
df[['house_num', 'street_addr']] = blocks.str.extract(MY_REGX.BLOCK, expand=True)
primary_type_post = [(None, capitalize_first)]
# Combine regex validation and max length 50
description_val = lambda series: series.str.match(MY_REGX.DESCRIPTION, na=False) & series.str.len().le(50)
description_post = [(None, capitalize_first)]
# Combine regex validation and max length 50
location_description_val = lambda series: series.str.match(MY_REGX.LOCATION_DESCRIPTION, na=False) & series.str.len().le(50)
location_description_post = [(None, capitalize_first)]
# arrest / domestic fields look to be all True / False, so let's just confirm this with a valid values constraint
tf_valid = ['true', 'false']
# beat, district, ward, community area, look to be all integer fields, so for these four, we will
# validate it's an int
valid_int = lambda series: series.str.isdigit()
# Location, we want to extract lat / lon into their own columns
# Also, we don't care about the original field after the extract, so we'll drop it
def location_lat_lon(df, locations):
df[['latitude','longitude']] = locations.str.extract(MY_REGX.LOCATION, expand=True)
# For zip codes we want to prefix 4 length zip codes with a '0' at the beginning after validation
def zip_to_five(zips):
condlist = [zips.str.len().eq(4), zips.str.len().eq(5)]
choicelist = ['0' + zips, zips]
return pd.Series(index=zips.index, data=np.select(condlist, choicelist, default=np.nan))
post_zip_codes = [(None, lambda series: zip_to_five(series))]
nullable_fields = {}
nullable_fields['block'] = {'validation': MY_REGX.BLOCK, 'generated_cols': [block_num_addr]}
nullable_fields['iucr'] = {'validation': MY_REGX.IUCR}
nullable_fields['primary_type'] = {'validation': MY_REGX.PRIMARY_TYPE, 'post_process': primary_type_post}
nullable_fields['description'] = {'validation': description_val, 'post_process': description_post}
nullable_fields['location_description'] = {'validation': location_description_val, 'post_process': location_description_post}
nullable_fields['arrest'] = {'valid_values': tf_valid}
nullable_fields['domestic'] = {'valid_values': tf_valid}
nullable_fields['beat'] = {'validation': valid_int}
nullable_fields['district'] = {'validation': valid_int}
nullable_fields['ward'] = {'validation': valid_int}
nullable_fields['community_area'] = {'validation': valid_int}
nullable_fields['location'] = {'validation': MY_REGX.LOCATION, 'generated_cols': [location_lat_lon], 'drop_field': True}
nullable_fields['zip_codes'] = {'validation': MY_REGX.ZIP_CODES, 'post_process': post_zip_codes}
for field, params in nullable_fields.items():
logger.debug(f'Processing {field}')
data_utils.process_field(df_filt, field, soft_rejects, **params)
return soft_rejects
# ## Now, let's test our scrubbing process on a single file
#
# First, we'll get a list of the files we want to process via Lambda
#
# <font color=red>Make sure to change the name of the S3_BUCKET to your bucket</font>
# +
s3 = s3fs.S3FileSystem()
FILE_PATTERN = re.compile('.*?(\d+)\.csv')
S3_BUCKET = 'chi-town-scrub-data'
s3_files = aws_utils.get_s3_files_to_process(s3, FILE_PATTERN, S3_BUCKET, '')
s3_files
# -
# ## Pick one of the files above to process
# +
# %%time
s3_bucket_key = s3_files[0][1]
print(f'Loading {s3_bucket_key}')
with s3.open(s3_files[0][1], 'r') as file:
df = pd.read_csv(file, low_memory=False, encoding='utf-8', dtype=str)
# I like column names lower case and with spaces replaced with underscores
data_utils.clean_col_names(df)
keep_cols = ['id', 'case_number', 'date', 'block', 'iucr', 'primary_type', 'description', 'location_description',
'arrest', 'domestic', 'beat', 'district', 'ward', 'community_area', 'location', 'zip_codes']
# Filter out the data to the columns we're interested in
df = df[keep_cols]
# This will trim data as well as replace multiple white spaces inside text with a single white space
data_utils.remove_excess_white_space(df)
# -
# ### We're not focused too much in this example on exploring the data, but it's useful to take a quick peak at the data in the columns to get an idea of it, and to help visualize what the regex is doing
df.head()
# +
print(f'Number of crimes: {len(df):,}')
print(f'Number of crimes resulting in arrest: {len(df.arrest.eq("true")):,}')
print('\nMost common crimes (primary type):\n')
crime_cnt = Counter(df.primary_type)
for primary_type, cnt in crime_cnt.most_common(5):
print(f'{primary_type} => {cnt:,}')
crime_cnt_spec = Counter(df['primary_type'] + ' => ' + df['description'])
print('\nMost common crimes (primary type => description):\n')
for specific_type, cnt in crime_cnt_spec.most_common(10):
print(f'{specific_type} => {cnt:,}')
crime_loc = Counter(df.location_description)
print('\nMost common crime location:\n')
for location, cnt in crime_loc.most_common(10):
print(f'{location} => {cnt:,}')
# -
# ## Now let's clean the data
#
# This is essentially what we want the Lambda function to do for us
#
# 1. Process hard rejects (and filter our DataFrame)
# 2. Process soft rejects
# 3. Analyze and log the rejects (so we know where to focus improvements in our scrubbing regex)
# 4. Upload the reject data to s3, as well as our clean data
# +
# %%time
df_filt, hard_rejects = process_hard_rejects(df)
soft_rejects = process_soft_rejects(df_filt)
unq_hard_rejects, unq_soft_rejects = data_utils.analyze_hard_and_soft_rejects(hard_rejects, soft_rejects)
file_name = s3_bucket_key.split('/')[-1]
hard_rej_df, soft_rej_df = data_utils.generate_reject_dfs(df, df_filt, file_name, hard_rejects, unq_hard_rejects, soft_rejects, unq_soft_rejects)
with s3.open(f'{S3_BUCKET}/rejects/hard_rejects_{file_name}', 'w') as hard_up, \
s3.open(f'{S3_BUCKET}/rejects/soft_rejects_{file_name}', 'w') as soft_up, \
s3.open(f'{S3_BUCKET}/clean_data/clean_{file_name}', 'w') as clean_up:
hard_rej_df.to_csv(hard_up, index_label='file_index', encoding='utf-8')
soft_rej_df.to_csv(soft_up, index_label='file_index', encoding='utf-8')
clean_cols = [col for col in df_filt.columns if '_orig' not in col]
df_filt[clean_cols].to_csv(clean_up, index=False, encoding='utf-8')
# -
# <font color=blue>You can go check your S3 bucket now to see the data is there</font>
# ## Let's take a peek at our hard rejcts and soft rejects
#
# You will probably be looking these, tweaking your process, and re-running the above code over and over until your happy
# with the results, then set up your Lambda function to do the heavy lifting
#
# Notice that we are storing the file_name of the rejects, and that we uploaded this to s3. We can easily list all the rejects
# files in the rejects bucket we made in S3, load them all and concatenate the results to a single DataFrame and analyze everything
# after Lambda does it's work
#
# We are also storing the column(s) that caused the reject
hard_rej_df.head()
# ### For example, we stated as a hard reject validation in our regex that all "valid" case numbers, start with two letters, so let's check our rejects
hard_rej_df.loc[hard_rej_df.cols.str.contains('case_number'), 'case_number'].str.slice(0,2).unique()
# And for the "clean" data
df_filt['case_number'].str.slice(0,2).unique()
# ## Soft rejects, as they are not totally excluded, include the original value, so you can see what is being set to null
soft_rej_df.head()
# ### Similar to the function we used above, analyze_hard_and_soft_rejects, we can look at them this way
orig_cols = [col for col in soft_rej_df.columns if '_orig' in col]
soft_rej_df[orig_cols].notnull().sum()
# ### We can see what's being set to null and then tweak our regex
#
# Here, it looks like our description validation is probably too strict, so there's room for improvement
MY_REGX.DESCRIPTION
soft_rej_df.loc[soft_rej_df.description_orig.notnull(), ['description', 'description_orig']].sample(20)
# ### It's also useful to look at the "clean" DataFrame, and sample it for the columns you need to clean up and see if anything weird is still in there and tweak your regex
df_filt[clean_cols].sample(10)
# ## Now let's turn this into a Lambda!
jb25zdCBzdGVwcyA9IG91dHB1dEVsZW1lbnQuc3RlcHM7CgogIGNvbnN0IG5leHQgPSBzdGVwcy5uZXh0KG91dHB1dEVsZW1lbnQubGFzdFByb21pc2VWYWx1ZSk7CiAgcmV0dXJuIFByb21pc2UucmVzb2x2ZShuZXh0LnZhbHVlLnByb21pc2UpLnRoZW4oKHZhbHVlKSA9PiB7CiAgICAvLyBDYWNoZSB0aGUgbGFzdCBwcm9taXNlIHZhbHVlIHRvIG1ha2UgaXQgYXZhaWxhYmxlIHRvIHRoZSBuZXh0CiAgICAvLyBzdGVwIG9mIHRoZSBnZW5lcmF0b3IuCiAgICBvdXRwdXRFbGVtZW50Lmxhc3RQcm9taXNlVmFsdWUgPSB2YWx1ZTsKICAgIHJldHVybiBuZXh0LnZhbHVlLnJlc3BvbnNlOwogIH0pOwp9CgovKioKICogR2VuZXJhdG9yIGZ1bmN0aW9uIHdoaWNoIGlzIGNhbGxlZCBiZXR3ZWVuIGVhY2ggYXN5bmMgc3RlcCBvZiB0aGUgdXBsb2FkCiAqIHByb2Nlc3MuCiAqIEBwYXJhbSB7c3RyaW5nfSBpbnB1dElkIEVsZW1lbnQgSUQgb2YgdGhlIGlucHV0IGZpbGUgcGlja2VyIGVsZW1lbnQuCiAqIEBwYXJhbSB7c3RyaW5nfSBvdXRwdXRJZCBFbGVtZW50IElEIG9mIHRoZSBvdXRwdXQgZGlzcGxheS4KICogQHJldHVybiB7IUl0ZXJhYmxlPCFPYmplY3Q+fSBJdGVyYWJsZSBvZiBuZXh0IHN0ZXBzLgogKi8KZnVuY3Rpb24qIHVwbG9hZEZpbGVzU3RlcChpbnB1dElkLCBvdXRwdXRJZCkgewogIGNvbnN0IGlucHV0RWxlbWVudCA9IGRvY3VtZW50LmdldEVsZW1lbnRCeUlkKGlucHV0SWQpOwogIGlucHV0RWxlbWVudC5kaXNhYmxlZCA9IGZhbHNlOwoKICBjb25zdCBvdXRwdXRFbGVtZW50ID0gZG9jdW1lbnQuZ2V0RWxlbWVudEJ5SWQob3V0cHV0SWQpOwogIG91dHB1dEVsZW1lbnQuaW5uZXJIVE1MID0gJyc7CgogIGNvbnN0IHBpY2tlZFByb21pc2UgPSBuZXcgUHJvbWlzZSgocmVzb2x2ZSkgPT4gewogICAgaW5wdXRFbGVtZW50LmFkZEV2ZW50TGlzdGVuZXIoJ2NoYW5nZScsIChlKSA9PiB7CiAgICAgIHJlc29sdmUoZS50YXJnZXQuZmlsZXMpOwogICAgfSk7CiAgfSk7CgogIGNvbnN0IGNhbmNlbCA9IGRvY3VtZW50LmNyZWF0ZUVsZW1lbnQoJ2J1dHRvbicpOwogIGlucHV0RWxlbWVudC5wYXJlbnRFbGVtZW50LmFwcGVuZENoaWxkKGNhbmNlbCk7CiAgY2FuY2VsLnRleHRDb250ZW50ID0gJ0NhbmNlbCB1cGxvYWQnOwogIGNvbnN0IGNhbmNlbFByb21pc2UgPSBuZXcgUHJvbWlzZSgocmVzb2x2ZSkgPT4gewogICAgY2FuY2VsLm9uY2xpY2sgPSAoKSA9PiB7CiAgICAgIHJlc29sdmUobnVsbCk7CiAgICB9OwogIH0pOwoKICAvLyBXYWl0IGZvciB0aGUgdXNlciB0byBwaWNrIHRoZSBmaWxlcy4KICBjb25zdCBmaWxlcyA9IHlpZWxkIHsKICAgIHByb21pc2U6IFByb21pc2UucmFjZShbcGlja2VkUHJvbWlzZSwgY2FuY2VsUHJvbWlzZV0pLAogICAgcmVzcG9uc2U6IHsKICAgICAgYWN0aW9uOiAnc3RhcnRpbmcnLAogICAgfQogIH07CgogIGNhbmNlbC5yZW1vdmUoKTsKCiAgLy8gRGlzYWJsZSB0aGUgaW5wdXQgZWxlbWVudCBzaW5jZSBmdXJ0aGVyIHBpY2tzIGFyZSBub3QgYWxsb3dlZC4KICBpbnB1dEVsZW1lbnQuZGlzYWJsZWQgPSB0cnVlOwoKICBpZiAoIWZpbGVzKSB7CiAgICByZXR1cm4gewogICAgICByZXNwb25zZTogewogICAgICAgIGFjdGlvbjogJ2NvbXBsZXRlJywKICAgICAgfQogICAgfTsKICB9CgogIGZvciAoY29uc3QgZmlsZSBvZiBmaWxlcykgewogICAgY29uc3QgbGkgPSBkb2N1bWVudC5jcmVhdGVFbGVtZW50KCdsaScpOwogICAgbGkuYXBwZW5kKHNwYW4oZmlsZS5uYW1lLCB7Zm9udFdlaWdodDogJ2JvbGQnfSkpOwogICAgbGkuYXBwZW5kKHNwYW4oCiAgICAgICAgYCgke2ZpbGUudHlwZSB8fCAnbi9hJ30pIC0gJHtmaWxlLnNpemV9IGJ5dGVzLCBgICsKICAgICAgICBgbGFzdCBtb2RpZmllZDogJHsKICAgICAgICAgICAgZmlsZS5sYXN0TW9kaWZpZWREYXRlID8gZmlsZS5sYXN0TW9kaWZpZWREYXRlLnRvTG9jYWxlRGF0ZVN0cmluZygpIDoKICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgJ24vYSd9IC0gYCkpOwogICAgY29uc3QgcGVyY2VudCA9IHNwYW4oJzAlIGRvbmUnKTsKICAgIGxpLmFwcGVuZENoaWxkKHBlcmNlbnQpOwoKICAgIG91dHB1dEVsZW1lbnQuYXBwZW5kQ2hpbGQobGkpOwoKICAgIGNvbnN0IGZpbGVEYXRhUHJvbWlzZSA9IG5ldyBQcm9taXNlKChyZXNvbHZlKSA9PiB7CiAgICAgIGNvbnN0IHJlYWRlciA9IG5ldyBGaWxlUmVhZGVyKCk7CiAgICAgIHJlYWRlci5vbmxvYWQgPSAoZSkgPT4gewogICAgICAgIHJlc29sdmUoZS50YXJnZXQucmVzdWx0KTsKICAgICAgfTsKICAgICAgcmVhZGVyLnJlYWRBc0FycmF5QnVmZmVyKGZpbGUpOwogICAgfSk7CiAgICAvLyBXYWl0IGZvciB0aGUgZGF0YSB0byBiZSByZWFkeS4KICAgIGxldCBmaWxlRGF0YSA9IHlpZWxkIHsKICAgICAgcHJvbWlzZTogZmlsZURhdGFQcm9taXNlLAogICAgICByZXNwb25zZTogewogICAgICAgIGFjdGlvbjogJ2NvbnRpbnVlJywKICAgICAgfQogICAgfTsKCiAgICAvLyBVc2UgYSBjaHVua2VkIHNlbmRpbmcgdG8gYXZvaWQgbWVzc2FnZSBzaXplIGxpbWl0cy4gU2VlIGIvNjIxMTU2NjAuCiAgICBsZXQgcG9zaXRpb24gPSAwOwogICAgZG8gewogICAgICBjb25zdCBsZW5ndGggPSBNYXRoLm1pbihmaWxlRGF0YS5ieXRlTGVuZ3RoIC0gcG9zaXRpb24sIE1BWF9QQVlMT0FEX1NJWkUpOwogICAgICBjb25zdCBjaHVuayA9IG5ldyBVaW50OEFycmF5KGZpbGVEYXRhLCBwb3NpdGlvbiwgbGVuZ3RoKTsKICAgICAgcG9zaXRpb24gKz0gbGVuZ3RoOwoKICAgICAgY29uc3QgYmFzZTY0ID0gYnRvYShTdHJpbmcuZnJvbUNoYXJDb2RlLmFwcGx5KG51bGwsIGNodW5rKSk7CiAgICAgIHlpZWxkIHsKICAgICAgICByZXNwb25zZTogewogICAgICAgICAgYWN0aW9uOiAnYXBwZW5kJywKICAgICAgICAgIGZpbGU6IGZpbGUubmFtZSwKICAgICAgICAgIGRhdGE6IGJhc2U2NCwKICAgICAgICB9LAogICAgICB9OwoKICAgICAgbGV0IHBlcmNlbnREb25lID0gZmlsZURhdGEuYnl0ZUxlbmd0aCA9PT0gMCA/CiAgICAgICAgICAxMDAgOgogICAgICAgICAgTWF0aC5yb3VuZCgocG9zaXRpb24gLyBmaWxlRGF0YS5ieXRlTGVuZ3RoKSAqIDEwMCk7CiAgICAgIHBlcmNlbnQudGV4dENvbnRlbnQgPSBgJHtwZXJjZW50RG9uZX0lIGRvbmVgOwoKICAgIH0gd2hpbGUgKHBvc2l0aW9uIDwgZmlsZURhdGEuYnl0ZUxlbmd0aCk7CiAgfQoKICAvLyBBbGwgZG9uZS4KICB5aWVsZCB7CiAgICByZXNwb25zZTogewogICAgICBhY3Rpb246ICdjb21wbGV0ZScsCiAgICB9CiAgfTsKfQoKc2NvcGUuZ29vZ2xlID0gc2NvcGUuZ29vZ2xlIHx8IHt9OwpzY29wZS5nb29nbGUuY29sYWIgPSBzY29wZS5nb29nbGUuY29sYWIgfHwge307CnNjb3BlLmdvb2dsZS5jb2xhYi5fZmlsZXMgPSB7CiAgX3VwbG9hZEZpbGVzLAogIF91cGxvYWRGaWxlc0NvbnRpbnVlLAp9Owp9KShzZWxmKTsK", "ok": true, "headers": [["content-type", "application/javascript"]], "status": 200, "status_text": "OK"}}, "base_uri": "https://localhost:8080/", "height": 163} id="ZsIVDYhkpRLd" outputId="6f92a530-e180-4a8b-db7e-21a7bae1dd99"
from google.colab import files
uploaded = files.upload()
for fn in uploaded.keys():
print('User uploaded file "{name}" with length {length} bytes'.format(
name=fn, length=len(uploaded[fn])))
# + id="gRGCStv2pohW"
embeddings = get_embeddings(uploaded)
# + colab={"base_uri": "https://localhost:8080/"} id="gvBdrY9astFI" outputId="42690ce9-7f70-4b00-c9b5-1c8b47d70019"
print(embeddings)
# + colab={"base_uri": "https://localhost:8080/"} id="d2Iblj-vsu5b" outputId="1c8fd5eb-0b5a-40e5-a3e2-5bd37d4d5ba4"
is_match(embeddings[0], embeddings[1])
# + colab={"base_uri": "https://localhost:8080/"} id="W2Nh1HIYs2v4" outputId="91c95389-f4d5-42b2-fe92-0381f0cff93e"
is_match(embeddings[0], embeddings[2])
# + colab={"base_uri": "https://localhost:8080/"} id="WDs5nMPis46H" outputId="72386439-7025-4611-dfae-838ff99d37b4"
is_match(embeddings[1], embeddings[1])
# + colab={"base_uri": "https://localhost:8080/"} id="ok_g8cF0117O" outputId="7224462c-7046-486e-c19a-cf94240f9166"
embeddings.shape
| 19,625 |
/lesson_5/AdditionalMaterials/.ipynb_checkpoints/Ch5- Sentence Classification with CNN-checkpoint.ipynb | 2b93b524ba7797c58505755d9e16f661adbb7bce | [] | no_license | NickMandylas/deep-learning-lessons | https://github.com/NickMandylas/deep-learning-lessons | 1 | 0 | null | null | null | null | Jupyter Notebook | false | false | .py | 41,412 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# %matplotlib inline
from __future__ import print_function
import collections
import math
import numpy as np
import os
import random
import tensorflow as tf
import zipfile
from matplotlib import pylab
from six.moves import range
from six.moves.urllib.request import urlretrieve
import tensorflow as tf
from sklearn import preprocessing
from sklearn.cross_validation import train_test_split
class DataManager:
def __init__(self, verbose=True):
self.verbose= verbose
self.max_sentence_len= 0
self.questions= list()
self.str_labels= list()
self.numeral_labels= list()
self.numeral_data= list()
self.cur_pos=0
def maybe_download(self, dir_name, file_name, url):
if not os.path.exists(dir_name):
os.mkdir(dir_name)
if not os.path.exists(os.path.join(dir_name, file_name)):
urlretrieve(url + file_name, os.path.join(dir_name, file_name))
if self.verbose:
print("Downloaded successfully {}".format(file_name))
def read_data(self, dir_name, file_name):
file_path= os.path.join(dir_name, file_name)
self.questions= list(); self.labels= list()
with open(file_path, "r", encoding="latin-1") as f:
for row in f:
row_str= row.split(":")
label, question= row_str[0], row_str[1]
question= question.lower()
self.labels.append(label)
self.questions.append(question.split())
if self.max_sentence_len < len(self.questions[-1]):
self.max_sentence_len= len(self.questions[-1])
le= preprocessing.LabelEncoder()
le.fit(self.labels)
self.numeral_labels = le.transform(self.labels)
self.str_classes= le.classes_
self.num_classes= len(self.str_classes)
if self.verbose:
print("Sample questions \n")
print(self.questions[0:5])
print("Labels {}\n\n".format(self.str_classes))
def padding(self, length):
for question in self.questions:
question= question.extend(["pad"]*(length- len(question)))
def build_numeral_data(self, dictionary):
self.numeral_data= list()
for question in self.questions:
data= list()
for word in question:
data.append(dictionary[word])
self.numeral_data.append(data)
if self.verbose:
print('Sample numeral data \n')
print(self.numeral_data[0:5])
def train_valid_split(self, train_size=0.9, rand_seed=33):
X_train, X_valid, y_train, y_valid = train_test_split(np.array(self.numeral_data), np.array(self.numeral_labels),
test_size = 1-train_size, random_state= rand_seed)
self.train_numeral= X_train
self.train_labels= y_train
self.valid_numeral= X_valid
self.valid_labels= y_valid
@staticmethod
def build_dictionary_count(questions):
count= []
dictionary= dict()
words= []
for question in questions:
words.extend(question)
count.extend(collections.Counter(words).most_common())
for word,freq in count:
dictionary[word]= len(dictionary)
reverse_dictionary= dict(zip(dictionary.values(), dictionary.keys()))
return dictionary, reverse_dictionary, count
def next_batch(self, batch_size, vocab_size, input_len):
data_batch= np.zeros([batch_size, input_len, vocab_size])
label_batch= np.zeros([batch_size, self.num_classes])
train_size= len(self.train_numeral)
for i in range(batch_size):
for j in range(input_len):
data_batch[i,j, self.train_numeral[self.cur_pos][j]]=1
label_batch[i, self.train_labels[self.cur_pos]]=1
self.cur_pos= (self.cur_pos+1)%train_size
return data_batch, label_batch
def convert_to_feed(self, data_numeral, label_numeral, input_len, vocab_size):
data2feed= np.zeros([data_numeral.shape[0], input_len, vocab_size])
label2feed= np.zeros([data_numeral.shape[0], self.num_classes])
for i in range(data_numeral.shape[0]):
for j in range(input_len):
data2feed[i,j, data_numeral[i][j]]=1
label2feed[i, label_numeral[i]]=1
return data2feed, label2feed
train_dm = DataManager()
train_dm.maybe_download("Data/question-classif-data", "train_1000.label", "http://cogcomp.org/Data/QA/QC/")
test_dm = DataManager()
test_dm.maybe_download("Data/question-classif-data", "TREC_10.label", "http://cogcomp.org/Data/QA/QC/")
train_dm.read_data("Data/question-classif-data", "train_1000.label")
test_dm.read_data("Data/question-classif-data", "TREC_10.label")
pad_len = max(train_dm.max_sentence_len, test_dm.max_sentence_len)
train_dm.padding(pad_len)
test_dm.padding(pad_len)
all_questions= list(train_dm.questions)
all_questions.extend(test_dm.questions)
dictionary,_,_= DataManager.build_dictionary_count(all_questions)
train_dm.build_numeral_data(dictionary)
test_dm.build_numeral_data(dictionary)
train_dm.train_valid_split()
data_batch, label_batch= train_dm.next_batch(batch_size=5, vocab_size= len(dictionary), input_len= pad_len)
print("Sample data batch- label batch \n")
print(data_batch)
print(label_batch)
class Layers:
@staticmethod
def dense(inputs, output_size, name="dense1", act=None):
with tf.name_scope(name):
input_size= int(inputs.get_shape()[1])
W_init = tf.random_normal([input_size, output_size], mean=0, stddev= 0.1, dtype= tf.float32)
b_init= tf.random_normal([output_size], mean=0, stddev= 0.1, dtype= tf.float32)
W= tf.Variable(W_init, name= "W")
b= tf.Variable(b_init, name="b")
Wxb= tf.matmul(inputs, W) + b
if act is None:
return Wxb
else:
return act(Wxb)
@staticmethod
def conv2D(inputs, filter_shape, strides=[1,1,1,1], padding="SAME", name= "conv1", act=None):
with tf.name_scope(name):
W_init= tf.random_normal(filter_shape, mean=0, stddev=0.1, dtype= tf.float32)
W= tf.Variable(W_init, name="W")
b_init= tf.random_normal([int(filter_shape[3])], mean=0, stddev=0.1, dtype= tf.float32)
b= tf.Variable(b_init, name="b")
Wxb= tf.nn.conv2d(input= inputs, filter= W, strides= strides, padding= padding)+b
if act is None:
return Wxb
else:
return act(Wxb)
@staticmethod
def conv1D(inputs, filter_shape, stride=1, padding="SAME", name="conv1", act=None):
with tf.name_scope(name):
W_init= tf.random_normal(filter_shape, mean=0, stddev=0.1, dtype= tf.float32)
W= tf.Variable(W_init, name="W")
b_init= tf.random_normal([filter_shape[2]], mean=0, stddev=0.1)
b= tf.Variable(b_init, name="b")
Wxb= tf.nn.conv1d(value=inputs, filters=W, stride= stride, padding= padding) +b
if act is None:
return Wxb
else:
return act(Wxb)
@staticmethod
def max_pool(inputs, ksize=[1,2,2,1],strides=[1,2,2,1], padding="SAME"):
return tf.nn.max_pool(value= inputs, ksize=ksize, strides= strides, padding= padding)
@staticmethod
def dropout(inputs, keep_prob):
return tf.nn.dropout(inputs, keep_prob= keep_prob)
@staticmethod
def batch_norm(inputs, phase_train):
return tf.contrib.layers.batch_norm(inputs, decay= 0.99,
is_training=phase_train, center= True, scale=True, reuse= False)
# +
class SC_CNN:
def __init__(self, height, width, batch_size=32, epochs=100, num_classes=5, save_history= True,
verbose= True, optimizer= tf.train.AdamOptimizer(learning_rate=0.001), learning_rate=0.001):
tf.reset_default_graph()
self.height= height
self.width= width
self.batch_size= batch_size
self.epochs= epochs
self.num_classes= num_classes
self.optimizer= optimizer
self.optimizer.learning_rate= learning_rate
self.verbose= verbose
self.save_history= save_history
if self.save_history:
self.H= {"train_loss_batch": [], "train_acc_batch": [], "train_loss_epoch": [],
"train_acc_epoch": [], "valid_loss_epoch": [], "valid_acc_epoch": []}
self.session= tf.Session()
def build(self):
self.X= tf.placeholder(shape= [None, self.height, self.width], dtype=tf.float32)
self.y= tf.placeholder(shape= [None, self.num_classes], dtype= tf.float32)
conv1= Layers.conv1D(inputs= self.X, filter_shape= [3, self.width, 1], name="conv1")
conv2= Layers.conv1D(inputs= self.X, filter_shape= [5, self.width, 1], name="conv2")
conv3= Layers.conv1D(inputs= self.X, filter_shape= [7, self.width, 1], name="conv3")
h1=tf.reduce_max(conv1, axis=1)
h2=tf.reduce_max(conv2, axis=1)
h3= tf.reduce_max(conv3, axis=1)
h= tf.concat([h1,h2,h3], axis=1)
logits= Layers.dense(inputs= h, output_size= self.num_classes)
with tf.name_scope("train"):
cross_entropy= tf.nn.softmax_cross_entropy_with_logits(labels=self.y, logits= logits)
self.loss= tf.reduce_mean(cross_entropy)
self.train= self.optimizer.minimize(self.loss)
with tf.name_scope("predict"):
self.y_pred= tf.argmax(logits, axis=1)
y1= tf.argmax(self.y, axis=1)
corrections= tf.cast(tf.equal(self.y_pred, y1), dtype=tf.float32)
self.accuracy= tf.reduce_mean(corrections)
self.session.run(tf.global_variables_initializer())
def partial_fit(self, data_batch, label_batch):
self.session.run([self.train], feed_dict={self.X:data_batch, self.y:label_batch})
if self.save_history:
self.compute_loss_acc(data_batch, label_batch, "train", "Iteration", 1)
def predict(self, X,y):
y_pred, acc= self.session.run([self.y_pred, self.accuracy], feed_dict={self.X:X, self.y:y})
return y_pred, acc
def compute_loss_acc(self, X, y, applied_set="train", applied_scope="Epoch", index= 1):
loss, acc= self.session.run([self.loss, self.accuracy], feed_dict={self.X:X, self.y:y})
if self.verbose and applied_scope=="Epoch":
print("{} {} {} loss= {}, acc={}".format(applied_scope, index, applied_set, loss, acc))
if self.save_history:
if applied_scope=="Iteration":
self.H["train_loss_batch"].append(loss)
self.H["train_acc_batch"].append(acc)
else:
if applied_set=="train":
self.H["train_loss_epoch"].append(loss)
self.H["train_acc_epoch"].append(acc)
else:
self.H["valid_loss_epoch"].append(loss)
self.H["valid_acc_epoch"].append(acc)
# +
batch_size= 32
epochs= 100
train_size= len(train_dm.train_numeral)
iter_per_epoch= math.ceil(train_size/batch_size)
network= SC_CNN(height= pad_len, width= len(dictionary),batch_size=batch_size, epochs= epochs, num_classes= train_dm.num_classes)
network.build()
train2feed, train_label2feed= train_dm.convert_to_feed(train_dm.train_numeral, train_dm.train_labels,
input_len= pad_len, vocab_size=len(dictionary))
valid2feed, valid_label2feed= train_dm.convert_to_feed(train_dm.valid_numeral, train_dm.valid_labels,
input_len= pad_len, vocab_size=len(dictionary))
test2feed, test_label2feed= test_dm.convert_to_feed(np.array(test_dm.numeral_data), np.array(test_dm.numeral_labels),
input_len= pad_len, vocab_size=len(dictionary))
for epoch in range(epochs):
for i in range(iter_per_epoch):
data_batch, label_batch= train_dm.next_batch(batch_size= batch_size,
vocab_size=len(dictionary), input_len= pad_len)
#print(data_batch.shape, label_batch.shape)
network.partial_fit(data_batch, label_batch)
network.compute_loss_acc(train2feed, train_label2feed, "train", "Epoch", epoch +1)
network.compute_loss_acc(valid2feed, valid_label2feed, "valid", "Epoch", epoch +1)
print("Finish training and computing testing performance\n")
y_pred, test_acc= network.predict(test2feed, test_label2feed)
print("Testing accuracy: {}".format(test_acc))
# +
import matplotlib.pyplot as plt
# %matplotlib inline
plt.rcParams["figure.figsize"] = (20,3)
def plot_history(history):
plt.subplot(2,1,1)
plt.plot(history["train_loss_epoch"], "r^-", label="train loss epoch")
plt.plot(history["valid_loss_epoch"], "b*-", label= "valid loss epoch")
plt.legend()
plt.subplot(2,1,2)
plt.plot(history["train_acc_epoch"], "r^-", label="train acc epoch")
plt.plot(history["valid_acc_epoch"], "b*-", label= "valid acc epoch")
plt.legend()
plt.show()
plot_history(network.H)
# -
| 13,613 |
/labs/Lab00.ipynb | 3576cef767181b884327c5d7fb78f96a3c2013fa | [] | no_license | ozencgungor/p250-spring-2021 | https://github.com/ozencgungor/p250-spring-2021 | 0 | 0 | null | null | null | null | Jupyter Notebook | false | false | .py | 22,721 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Lab 00 : An Introduction
# ### Objectives
#
# This lab is "extra credit". Its main purpose is to get us working with the tools, working as a group, and ironing out all the details in the process. The same procedure will be used for all the labs and for the computational portion of exams! All group members must be able to use the system on their own! In more detail, the main objectives are
#
# - Ensure that we have the working, correct version of Python and the Jupyter system installed.
# - Develop a work flow for the labs, including working with your group, submitting the lab, etc.
# - Learn some basics of plotting.
# - Learn some basic functions.
# ## Structure Overview
#
# The labs and prelabs will be structured in similar ways. There are text cells and code cells.
#
# The text cells (such as this one) will contain information for you to read or a place for you provide a response. Any text cell that contains
# ```
# YOUR ANSWER HERE
# ```
# is a cell that you must edit and provide your response to a question. You should delete the existing text and replace it with your response.
#
# The code cells may already contain code (such as the first code cell below). You should run this cell as provided. In addition it may also contain the code
# ```
# # YOUR CODE HERE
# raise NotImplementedError()
# ```
# This is a placeholder that will raise an error. You should delete these two lines and replace them with code you have written.
# ## Versions
#
# As stated in class and on the course website, you **must** be using `Python` version 3.6 (or later) and `IPython` version 6.0 (or later). If you are using the Anaconda distribution, as suggested on the course website, *and* you downloaded the correct version, you should be fine.
#
# Run the following cell to determine what versions of the software you are using. If there are any errors or if you do not have the correct versions, this must be corrected before proceding.
# +
import sys
import IPython
import numpy
import scipy
import matplotlib
print("Python version:", sys.version)
print("IPython version:", IPython.__version__)
print("NumPy version:", numpy.__version__)
print("SciPy version:", scipy.__version__)
print("Matplotlib version:", matplotlib.__version__)
# -
# ## Down with pylab!
#
# As a warning, there is an old fashioned way of doing plotting using the so called `pylab` interface. This is one of the original ways of creating plots and is heavily modelled after `Matlab`. Not surprisingly, it is a very poor interface and should **never be used**. It can be invoked in a couple of ways. I will mention them here so that if you ever see them being used in something you read online, code given to you, *etc.*, immediately stop reading it and run away as fast as you can!
#
# The things to watch out for are
# - `%pylab`
# - `from matplotlib.pyplot import *`
#
# **Never, never, never** use either of these.
# ## Plotting: Pyplot interface
#
# The simplest interface for "quick plots" is the `pyplot` interface. This is not what we will use for most of our plotting when we are creating quality figures, but it is quite useful when we want to quickly look at things.
#
# We will learn much more about this throughout the semester to the point that the following code will become second nature. For now, this can be used as boilerplate. Run the following cell to set up the numerical python and plotting environment for us.
import numpy as np
import matplotlib.pyplot as plt
# %matplotlib inline
# We are now ready to produce our first plot. Here we will make a histogram of 10,000 Gaussian random values. The code below does this for us.
#
# At this point you should not worry about the details of code that is provided. We will learn about these as we go along. On the other hand, it should make sense. A Gaussian distribution is also called the normal distribution, hence ....
#
# Run the following cell.
plt.hist(np.random.normal(size=10000), 1000)
# What *should* have happened is that a long list of numbers and some other information is printed and after this a figure with the histogram in it. The fact that the figure appears in the notebook is due to us using `inline` in our call to `%matplotlib` above. We prefer for our notebooks to be self contained, that is, anyone can open them up and see all the results without having to run any of the cells themselves. Hence we will always inline our figures.
#
# While this is good, printing all those numbers was not useful. It is nice to have access to this information, but looking at it just clutters the notebook, we just wanted the figure!
#
# To **not** get the output from a function (the long list of numbers above) we can use another feature of the notebook: a semicolon at the end of a line suppresses output. Note that this is a feature of the notebook, not a feature of the `Python` language. Run the following cell. This should produce a nice histogram of a Gaussian in the notebook itself without all the extra output we saw above.
plt.hist(np.random.normal(size=10000), 1000);
# ## Some Available Functions
#
# For most things we want to do, there are existing functions that will make our life much easier. For numerical work, `numpy` contains most of the basic utilities. When we run
# ```
# import numpy as np
# ```
# we load the `numpy` module and give it the short name (alias) `np`. We can access functions in the `numpy` namespace using `np`, which is shorter to type and a standard convention. You will find most code that uses `numpy` written with this shortcut.
# ### `arange`
#
# The `arange` function, as the name suggests, is used to generate a range of numbers (as a `numpy` array, this fact will be more useful and meaningful in the future). Here we will explore this function. Recall that we need to access this function (and every other `numpy` function) by prepending `np`. This means that we access this function as `np.arange`.
#
# Check the documentation for `arange`. You should see that there are two optional arguments, `start` and `step` (this is what is meant when the arguments are listed in square brackets) and that `dtype` has a default value (which we will ignore for now).
# ##### Question
#
# What are the default values for `start` and `step`?
# + [markdown] deletable=false nbgrader={"cell_type": "markdown", "checksum": "cfa75ff26b32271719f3267e51b19b83", "grade": true, "grade_id": "cell-30afaab95e8ea572", "locked": false, "points": 0.5, "schema_version": 3, "solution": true}
# YOUR ANSWER HERE
# -
# ##### Testing:
#
# The best way to get used to a function is to play around with it a bit. You should do that now. For your own edification, see what happens when you run `arange(0)`, `arange(2,0)`, `arange(3,0,-1)`. You should understand why these behave the way they do. (Again remember that we must always prepend `np.` to any function from the `numpy` module. You will not be warned of this again!)
#
# For these tests you can/should create code cell(s) below this one. Cells can be created using the "Insert" pull down menu or keyboard shortcuts. (One cell has been created for your convenience. Feel free to create more or just run a few tests in this cell. Make sure you leave one test in this cell for grading!)
# + deletable=false nbgrader={"cell_type": "code", "checksum": "bc4d84f6110d224992c7ad59d7be4d7e", "grade": true, "grade_id": "cell-5f14ce6cad82d450", "locked": false, "points": 0.5, "schema_version": 3, "solution": true}
# YOUR CODE HERE
raise NotImplementedError()
# -
# ###### Question:
#
# Show how to use `arange` to generate a list of numbers from -4 to 4 in steps of 0.1.
# + deletable=false nbgrader={"cell_type": "code", "checksum": "a9955b92c229b63ed2d3a47190a5f41b", "grade": true, "grade_id": "cell-2930654011687055", "locked": false, "points": 0.5, "schema_version": 3, "solution": true}
# YOUR CODE HERE
raise NotImplementedError()
# -
# Did the generated range contain the endpoint, 4? Should it have? Sometimes we want it to, sometimes we do not. By default `arange` **does not contain the endpoint**.
#
# ##### Question:
#
# Modify your answer to the previous part to include the endpoint, 4.
# + deletable=false nbgrader={"cell_type": "code", "checksum": "219769c4c2c9204019edd21b53538e40", "grade": true, "grade_id": "cell-c99b0774c5cc2c0f", "locked": false, "points": 0.5, "schema_version": 3, "solution": true}
# YOUR CODE HERE
raise NotImplementedError()
# -
# ### `linspace`
#
# Also as noted in the `arange` documentation, an alternative function that serves a similar purpose is `linspace`. Look up the documentation for `linspace`. (In fact, whenever we encounter an unknown function we should look up its documentation!)
#
# ##### Question:
#
# Briefly compare and contrast the `linspace` and `arange` functions. Write a couple of sentences comparing them. (It is also worthwhile to redo the previous exploration of `arange` now with `linspace`.)
# + [markdown] deletable=false nbgrader={"cell_type": "markdown", "checksum": "4c30332786574828de21abe24af072b3", "grade": true, "grade_id": "cell-70a5c4c71e313b10", "locked": false, "points": 1, "schema_version": 3, "solution": true}
# YOUR ANSWER HERE
# -
# ## A "Real" Plot
#
# We will now take the first step toward creating a real plot. The default plots created by `matplotlib` are "ok", not great, but with a little effort we can make them very nice. We will work on this more throughout the semester.
#
# As an example, we will improve the plot we made above. The `random.normal` function is suppose to pick random values from a Gaussian distribution with zero mean and unit variance. We will test this by plotting the histogram along with the expected curve for a Gaussian distribution. To do so we will proceed as follows. All code should be put in the cell at the end of this section.
#
# ### Steps:
#
# 1. Generate 10,000 random numbers using `np.random.normal`. Store them in some variable (use whatever name you prefer).
# 2. Find the minimum and maximum values from your array. Looking through such a long array by hand would not be a good use of your time, so naturally there are functions for doing this. Find some appropriate functions and use them. Print the minimum and maximum values using the `print` function. (Note: There are a number of ways to do this, you only need to find one.)
# 3. When we generate a histogram using `hist` we can specify the binning in a few ways. In our original plot we told `hist` to use 1000 bins; it figured out how big to make them from the data we provided (and, in fact, returned this information). Alternatively, we can tell it what bins to use and we will do that here. Create an array of bin values in steps of 0.1 from some value smaller than the minimum in your array to some value larger than the maximum in your array. It makes sense to use "simple" numbers like 3.8 or 4.2 instead of an arbitrary real number. Store this array in some new variable.
# 4. Use the array from the previous part as the bins for your histogram. As always, check the documentation of `hist` to learn how to do this. (You will notice there is a `bins` keyword.)
# 5. We would now like to plot the *expected* Gaussian on top of the histogram. A general Gaussian is given by
# $$ f(x,\mu,\sigma) = \frac1{\sqrt{2\pi\sigma^2}} \exp\!\left[ -\frac{(x-\mu)^2}{2\sigma^2} \!\right], $$
# where $\mu$ is the mean and $\sigma$ is the variance (width) of the Gaussian. (What are the *expected* values for $\mu$ and $\sigma$ for the distribution used by `random.normal`?) Unfortunately, if you plot this function on top of the histogram, they will not lie on top of each other correctly. (You should try this!) The problem is that this function is normalized, but our histogram was not. This is easy to fix. Once again look at the documentation for `hist` and notice that it has a `density` keyword. (Note that there is also a `normed` keyword. As the documentation states, this is now deprecated and should not be used in new plots. There many other options that may be useful. For example, I prefer to make histograms as steps instead of being filled. Feel free to explore some of these options.)
# 6. We are now ready to plot the Gaussian. To do this use the `plot` function. When you look up its documentation you will find it has many, many options. It is good to play with some of them. The most useful ones for now are those that allow us to change the color, style, and width of the line. (Hint: We also need to *calculate* the Gaussian. This uses functions like exponentiation and square roots and a constant, $\pi$. Naturally there are functions defined to do these operations and the value of $\pi$ is also available to us. In what module that we have already loaded would you expect them to be in? What do you expect them to be called? Often things will work out as we expect.)
# 7. If you have performed the steps given above you now have a plot, but not a very useful plot. It contains a histogram and a line, but what are these things? What are we suppose to learn from the plot? There are a few things we should **always** do whenever we make a plot. We will have a more complete set of rules in the next lab but for now here are a few things to do to get us started:
# 1. **Required:** Label the axes. What is on the $x$ and $y$ axes in the plot? Without labels we have no idea. The labels on these axes should be descriptive so that at a quick glance we know what is being plotted. At this point you may be able to guess the names of the functions to use to label the axes. (Hint: Look up `xlabel`. Also, remember that any function that comes from a module must be prefixed by that modules name. For plotting we have chosen to use the alias `plt` for the `pyplot` interface.)
# 2. **Required:** Title the figure. For this course you must **always** add a title to your figures. When a figure is included in a paper you write it has a caption so a title is not necessary (nor should be included), but for this course you must always include one. It should come as no surprise what function to use to create a title!
# 3. **Suggested:** Add minor tick marks. By default only the major tick marks (with tick labels) are created. A figure almost always looks better with smaller ticks in-between the larger ones. These can be turned on using `minorticks_on()`.
# 8. Finally, put all of this together into a single code cell below that prints the minimum and maximum values and also produces the required plot.
# + deletable=false nbgrader={"cell_type": "code", "checksum": "53ed9a75cde94e70cd4f94610d7a2c08", "grade": true, "grade_id": "cell-17fa0d61fcfbcd7e", "locked": false, "points": 7, "schema_version": 3, "solution": true}
# YOUR CODE HERE
raise NotImplementedError()
# -
# ## Finishing Up
#
# We are **not** done yet! The end result of all of this work should be a single document, this notebook, that contains all the required work, documentation, and code. It should be a complete document that "works". When working on problems, we often try little things in cells, delete them, *etc.* Due to the way the notebook works, the backend kernel remembers all these intermediate steps (variables that were created/set, for example) even after we delete the cells containing the work. Thus we should verify that our document is complete. One way to do so is described below.
#
# ### Warning:
#
# Performing the following operation will erase all output and remove everything defined in your running kernel. If you do **not** have everything needed saved in cells in your current document then information will be lost. However, these steps **will not destroy anything you have entered and saved in the notebook**, it only erases the output and clears the state of the kernel.
#
# ### Operation:
#
# The simplest step is to use the *Kernel* pull-down menu and select *Restart & Run All*. This will pop up a warning telling you want it is going to do.
#
# ### Note:
#
# Your notebook **will be rerun before grading**. Even if you had left all the correct results/plots in your notebook when you uploaded it, if your notebook does not run, then it is incorrect. A notebook is a dynamical document, not a static one. Everyone (with the right environment installed) should be able to run it themselves and reproduce your results.
# ## Turning in the Lab
#
# Once everything is completed, the lab should be submitted by uploading it via canvas. For a group assignment, only one member of the group needs to submit the lab. Just make sure somebody does it!
#
# The **name of the notebook file cannot be changed**. The name of the uploaded file must be the same as the name of the file as listed in the assignment on canvas. Again, I will stress, **do not change the name of the file**. If the file is incorrectly named it will not be graded and you will receive no credit.
#
# For example, this file is called `Lab00.ipynb`. That is the name of the file that must be submitted. If this name got mangled in some way when you downloaded the file, rename it!
# ## Member Participation
#
# In the following cell enter the *Case ID* for each student in the group who partcipated in this lab. This is not just a list of group members, we already know who is in each group. Participation means having taken an active role in its development through discussions, coding, *etc.* By being included in this list you are affirming that each student is justified in claiming this lab as their work. Including someone who does not deserve credit is cheating. Such cases are a violation of academic integrity and will be treated as stated in the syllabus. If there are any questions or concerns regarding this, please contact the course instructor. All such communications will be treated with strict confidence.
# + [markdown] deletable=false nbgrader={"cell_type": "markdown", "checksum": "fff3c882c6eefc7c3dce10bb0b785d99", "grade": true, "grade_id": "cell-9d8dd31e7ce59c67", "locked": false, "points": 0, "schema_version": 3, "solution": true}
# YOUR ANSWER HERE
| 18,420 |
/factors_cohesion/social_cohesion_P3.ipynb | e2a42317df913b45bacdd3a3dc7c7bcdd529bf7c | [] | no_license | Tsedtopia/final_project | https://github.com/Tsedtopia/final_project | 1 | 0 | null | null | null | null | Jupyter Notebook | false | false | .py | 108,460 | # ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.15.2
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# +
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
from tqdm import tqdm
import seaborn as sns
import re
import string
from nltk.corpus import stopwords
from nltk.stem import SnowballStemmer
from string import punctuation
from nltk.stem.wordnet import WordNetLemmatizer
from sklearn.feature_extraction.text import TfidfVectorizer, CountVectorizer
from smart_open import open
from sklearn import preprocessing
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import MultiLabelBinarizer
from sklearn.naive_bayes import MultinomialNB
from sklearn.linear_model import SGDClassifier
from sklearn.linear_model import LogisticRegression
from sklearn import svm
from sklearn.metrics import f1_score
from sklearn import metrics
from sklearn.metrics import confusion_matrix
from sklearn.metrics import accuracy_score
from sklearn.metrics import classification_report
from sklearn.ensemble import RandomForestClassifier
from sklearn.metrics import mean_squared_error,confusion_matrix, precision_score, recall_score, auc,roc_curve
from sklearn.ensemble import GradientBoostingClassifier
from sklearn.ensemble import AdaBoostClassifier
from sklearn.pipeline import Pipeline
from sklearn.multiclass import OneVsRestClassifier
from sklearn.svm import LinearSVC
from sklearn.base import BaseEstimator
from sklearn.model_selection import GridSearchCV
from sklearn.discriminant_analysis import LinearDiscriminantAnalysis
import collections
from gensim.models.doc2vec import Doc2Vec, TaggedDocument
from nltk.tokenize import word_tokenize
from sklearn.decomposition import PCA
from sklearn.preprocessing import StandardScaler
import joblib
tqdm.pandas()
# %matplotlib inline
# -
train = pd.read_csv('../data/nlp_tweet_data/train.csv')
train.head()
train.shape
train.head()
test = pd.read_csv('../data/nlp_tweet_data/test.csv')
test.head()
test.shape
class_indexes = list(train['target'].value_counts().index)
all_data = train.append(test)
# ## Preprocessing
def clean_text(text, remove_stopwords=True, stem_words=False, lemma=True):
# Clean the text, with the option to remove stopwords and to stem words.
text = str(text).lower().split()
# Optionally, remove stop words
# if remove_stopwords:
# stops = set(stopwords.words("english"))
# text = [w for w in text if not w in stops]
text = " ".join(text)
# remove URLs
url = re.compile(r'https?://\S+|www\.\S+')
text = url.sub(r'',text)
# remove html tags
text = re.sub(r'<.*?>', "", text)
# remove emoji
emoji_pattern = re.compile("["
u"\U0001F600-\U0001F64F" # emoticons
u"\U0001F300-\U0001F5FF" # symbols & pictographs
u"\U0001F680-\U0001F6FF" # transport & map symbols
u"\U0001F1E0-\U0001F1FF" # flags (iOS)
u"\U00002702-\U000027B0"
u"\U000024C2-\U0001F251"
"]+", flags=re.UNICODE)
text = emoji_pattern.sub(r'', text)
# remove punctuation from text
text = "".join([char for char in text if char not in string.punctuation])
# Clean the text
text = re.sub(r"[-()\"#/<>!@&;*:<>{}`'+=~%|.!?,_]", " ", text)
text = re.sub(r"\]", " ", text)
text = re.sub(r"\[", " ", text)
text = re.sub(r"\/", " ", text)
text = re.sub(r"\\", " ", text)
text = re.sub(r"\'ve", " have ", text)
text = re.sub(r"can't", "cannot ", text)
text = re.sub(r"n't", " not ", text)
text = re.sub(r"\'re", " are ", text)
text = re.sub(r"\'d", " would ", text)
text = re.sub(r"\'ll", " will ", text)
text = re.sub(r" ", " ", text)
text = re.sub(r" ", " ", text)
text = re.sub(r" ", " ", text)
text = re.sub(r"0x00", "", text)
# Optionally, shorten words to their stems
if stem_words:
text = text.split()
stemmer = SnowballStemmer('english')
stemmed_words = [stemmer.stem(word) for word in text]
text = " ".join(stemmed_words)
if lemma:
text = text.split()
lem = WordNetLemmatizer()
lemmatized = [lem.lemmatize(word,"v") for word in text]
text = " ".join(lemmatized)
# Return a list of words
return(text)
all_data['text'] = all_data['text'].progress_apply(lambda x:clean_text(x))
train['text'] = train['text'].progress_apply(lambda x:clean_text(x))
test['text'] = test['text'].progress_apply(lambda x:clean_text(x))
# ## DOc2vec embedding
tagged_data = [TaggedDocument(words=word_tokenize(_d.lower()), tags=[str(i)]) for i, _d in enumerate(all_data['text'])]
tagged_data[1]
# ### Vocab building
# +
max_epochs = 100
vec_size = 100
alpha = 0.025
model = Doc2Vec(size=vec_size,
alpha=alpha,
min_alpha=0.00025,
min_count=1,
dm =1,
workers=12)
model.build_vocab(tagged_data)
# -
# ### Build Doc2vec model
# +
for epoch in range(max_epochs):
print('iteration {0}'.format(epoch))
model.train(tagged_data,
total_examples=model.corpus_count,
epochs=model.iter)
# decrease the learning rate
model.alpha -= 0.0002
# fix the learning rate, no decay
model.min_alpha = model.alpha
model.save("tweet_doc2vec100.model")
print("Model Saved")
# -
model= Doc2Vec.load("tweet_doc2vec100.model")
similar_doc = model.docvecs.most_similar('1')
print(similar_doc)
# ## convert doc2vec vectors to dataframe
data_list =[]
for i in range(len(model.docvecs)):
data_list.append(model.docvecs[i])
columns_name = [str(i) for i in range(len(model.docvecs[0]))]
d2v_df = pd.DataFrame.from_records(data_list,columns = columns_name)
d2v_df.head()
d2v_df.shape
# ## train test split
train_new=d2v_df[:train.shape[0]]
test_new=d2v_df[train.shape[0]:]
x_train, x_test, y_train, y_test = train_test_split(train_new, train['target'])
print('train shape: ',x_train.shape)
print('test shape: ',x_test.shape)
print("train_y label count: ",collections.Counter(y_train))
print("valid_y label count: ",collections.Counter(y_test))
# ## ML models
class MyClassifier(BaseEstimator):
def __init__(self, classifier_type):
"""
A Custome BaseEstimator that can switch between classifiers.
:param classifier_type: string - The switch for different classifiers
"""
self.classifier_type = classifier_type
def fit(self, X, y=None):
if self.classifier_type == 'LDA':
self.classifier_ = LinearDiscriminantAnalysis(solver='svd')
elif self.classifier_type == 'Logistic Regression':
self.classifier_ = LogisticRegression(C=1.0, dual= False, max_iter=500,penalty = 'l2' ,
solver = 'liblinear', class_weight='balanced',
multi_class='ovr', random_state=42, tol=1e-05)
elif self.classifier_type == 'LinSVC':
self.classifier_ = svm.LinearSVC(C=1.0, dual= False, fit_intercept=True,
intercept_scaling =0.1, max_iter=1000,penalty = 'l2' , loss = 'squared_hinge', class_weight='balanced',
multi_class='ovr', random_state=42, tol=1e-05, verbose=0)
elif self.classifier_type == 'Random Forest':
self.classifier_ = RandomForestClassifier(n_estimators = 10, criterion = 'entropy', random_state = 42)
elif self.classifier_type == 'Decision Tree':
self.classifier_ = DecisionTreeClassifier()
elif self.classifier_type == 'Gradient boost':
self.classifier_ = GradientBoostingClassifier(n_estimators=50)
else:
raise ValueError('Unkown classifier type.')
self.classifier_.fit(X, y)
return self
def predict(self, X, y=None):
return self.classifier_.predict(X)
def performance_plots(y_test_sample, y_pred, ml_algo):
print("---------"+ml_algo+"-----------")
acc = metrics.accuracy_score(y_pred, y_test_sample)
print("Accuracy: ",acc)
f1_val = f1_score(y_test_sample, y_pred)
print('f1 score:',f1_val)
print("NORMALISED CM for Tag6")
cm = confusion_matrix(y_test_sample,y_pred)
norm_cm = cm.astype('float') / cm.sum(axis=1)[:, np.newaxis]*100
ind = class_indexes
cols = class_indexes
cm_df = pd.DataFrame(norm_cm,cols,ind)
plt.figure(figsize=(22,10))
sns.heatmap(cm_df, annot=True,cmap = "Greens",fmt='g')
print(classification_report(y_test_sample,y_pred))
print(accuracy_score(y_test_sample, y_pred))
return
# ## LDA model
lda_model = MyClassifier('LDA')
lda_model.fit(x_train, y_train)
lda_pred = lda_model.predict(x_test)
performance_plots(y_test, lda_pred, 'LDA')
# ## SVM
svm_model = MyClassifier('LinSVC')
svm_model.fit(x_train, y_train)
svm_pred = svm_model.predict(x_test)
performance_plots(y_test, svm_pred, 'LinSVC')
el = 'Rolling Std')
plt.legend(loc='best')
plt.title('Rolling Mean & Standard Deviation')
plt.show(block=False)
dftest = adfuller(df)
# Extract and display test results in a user friendly manner
dfoutput = pd.Series(dftest[0:4], index=['Test Statistic', 'p-value', '#Lags Used', '# of Observations Used'])
for key,value in dftest[4].items():
dfoutput['Critical Value (%s)'%key] = value
print ('Results of Dickey-Fuller test: \n')
print(dfoutput)
stationarity_check(ts)
# # Perform a log and square root transform
# Plot a log transform of the original time series (ts).
# Plot a log transform
ts_log = np.log(ts)
fig = plt.figure(figsize=(12,6))
plt.plot(ts_log, color='blue');
# Plot a square root transform
ts_sqrt = np.sqrt(ts)
fig = plt.figure(figsize=(12,6))
plt.plot(ts_sqrt, color='blue');
# # Subtracting the rolling mean
# your code here
roll_mean = np.log(ts).rolling(window=7).mean()
fig = plt.figure(figsize=(11,7))
plt.plot(np.log(ts), color='blue',label='Original')
plt.plot(roll_mean, color='red', label='Rolling Mean')
plt.legend(loc='best')
plt.title('Rolling Mean & Standard Deviation')
plt.show(block=False)
# Subtract the moving average from the log transformed data
data_minus_roll_mean = np.log(ts) - roll_mean
data_minus_roll_mean.head(10)
# Drop the missing values
data_minus_roll_mean.dropna(inplace=True)
# Plot the result
fig = plt.figure(figsize=(11,7))
plt.plot(data_minus_roll_mean, color='red',label='Sales - rolling mean')
plt.legend(loc='best')
plt.title('Sales while the rolling mean is subtracted')
plt.show(block=False)
stationarity_check(data_minus_roll_mean)
"""
The time series are not stationary, as the p-value is still substantial
(0.15 instead of smaller than the typical threshold value 0.05).
"""
# # Subtracting the weighted rolling mean
#
# Repeat all the above steps to calculate the exponential weighted rolling mean with a halflife of 4. Start from the log-transformed data again. Compare the Dickey-Fuller test results. What do you conclude?
#
# +
# Calculate Weighted Moving Average of log transformed data
exp_roll_mean = np.log(ts).ewm(halflife=4).mean()
# Plot the original data with exp weighted average
fig = plt.figure(figsize=(12,7))
plt.plot(np.log(ts), color='blue',label='Original (Log Transformed)')
plt.plot(exp_roll_mean, color='red', label='Exponentially Weighted Rolling Mean')
plt.legend(loc='best')
plt.show(block=False)
# +
# Subtract this exponential weighted rolling mean from the log transformed data
# Print the resulting time series
# +
# Subtract the moving average from the original data and check head for Nans
data_minus_exp_roll_mean = np.log(ts) - exp_roll_mean
# Plot the time series
fig = plt.figure(figsize=(11,7))
plt.plot(data_minus_exp_roll_mean, color='blue',label='Passengers - weighted rolling mean')
plt.legend(loc='best')
plt.show(block=False)
# +
# Check for stationarity of data_minus_exp_roll_mean using your function.
# -
# Do a stationarity check
stationarity_check(data_minus_exp_roll_mean)
"""
The p-value of the Dickey-Fuller test <0.05, so this series seems to be stationary according to this test!
Do note that there is still strong seasonality.
"""
# # Differencing
# Using exponentially weighted moving averages, we seem to have removed the upward trend, but not the seasonality issue. Now use differencing to remove seasonality. Make sure you use the right amount of periods. Start from the log-transformed, exponentially weighted rolling mean-subtracted series.
#
# After you differenced the series, drop the missing values, plot the resulting time series, and then run the stationarity check() again.
# +
# Difference your data
data_diff = data_minus_exp_roll_mean.diff(periods=12)
# Drop the missing values
data_diff.dropna(inplace=True)
# Check out the first few rows
data_diff.head(5)
# -
# Plot your differenced time series
fig = plt.figure(figsize=(11,7))
plt.plot(data_diff, color='blue',label='passengers - rolling mean')
plt.legend(loc='best')
plt.title('Differenced passengers series')
plt.show(block=False)
# Perform the stationarity check
stationarity_check(data_diff)
"""
Even though the rolling mean and rolling average lines do seem to be fluctuating, the movements seem to be completely random,
and the same conclusion holds for the original time series. Your time series is now ready for modeling!
"""
| 13,858 |
Subsets and Splits