---
title: "CT-RATE Dataset"
license: cc-by-nc-sa-4.0
extra_gated_prompt: |
## Terms and Conditions for Using the CT-RATE Dataset
**1. Acceptance of Terms**
Accessing and using the CT-RATE dataset implies your agreement to these terms and conditions. If you disagree with any part, please refrain from using the dataset.
**2. Permitted Use**
- The dataset is intended solely for academic, research, and educational purposes.
- Any commercial exploitation of the dataset without prior permission is strictly forbidden.
- You must adhere to all relevant laws, regulations, and research ethics, including data privacy and protection standards.
**3. Data Protection and Privacy**
- Acknowledge the presence of sensitive information within the dataset and commit to maintaining data confidentiality.
- Direct attempts to re-identify individuals from the dataset are prohibited.
- Ensure compliance with data protection laws such as GDPR and HIPAA.
**4. Attribution**
- Cite the dataset and acknowledge the providers in any publications resulting from its use.
- Claims of ownership or exclusive rights over the dataset or derivatives are not permitted.
**5. Redistribution**
- Redistribution of the dataset or any portion thereof is not allowed.
- Sharing derived data must respect the privacy and confidentiality terms set forth.
**6. Disclaimer**
The dataset is provided "as is" without warranty of any kind, either expressed or implied, including but not limited to the accuracy or completeness of the data.
**7. Limitation of Liability**
Under no circumstances will the dataset providers be liable for any claims or damages resulting from your use of the dataset.
**8. Access Revocation**
Violation of these terms may result in the termination of your access to the dataset.
**9. Amendments**
The terms and conditions may be updated at any time; continued use of the dataset signifies acceptance of the new terms.
**10. Governing Law**
These terms are governed by the laws of the location of the dataset providers, excluding conflict of law rules.
**Consent:**
Accessing and using the CT-RATE dataset signifies your acknowledgment and agreement to these terms and conditions.
extra_gated_fields:
Name: "text"
Institution: "text"
Email: "text"
I have read and agree with Terms and Conditions for using the CT-RATE dataset: "checkbox"
configs:
- config_name: labels
data_files:
- split: train
path: "dataset/multi_abnormality_labels/train_predicted_labels.csv"
- split: validation
path: "dataset/multi_abnormality_labels/valid_predicted_labels.csv"
- config_name: reports
data_files:
- split: train
path: "dataset/radiology_text_reports/train_reports.csv"
- split: validation
path: "dataset/radiology_text_reports/validation_reports.csv"
- config_name: metadata
data_files:
- split: train
path: "dataset/metadata/train_metadata.csv"
- split: validation
path: "dataset/metadata/validation_metadata.csv"
---
# [Developing Generalist Foundation Models from a Multimodal Dataset for 3D Computed Tomography](https://arxiv.org/abs/2403.17834)
Welcome to the official page for [our paper](https://arxiv.org/abs/2403.17834), which introduces **CT-RATE**—a pioneering dataset in 3D medical imaging that uniquely pairs textual data with image data focused on chest CT volumes. Here, you will find the CT-RATE dataset, comprising chest CT volumes paired with corresponding radiology text reports, multi-abnormality labels, and metadata, all freely accessible to researchers.
## CT-RATE: A novel dataset of chest CT volumes with corresponding radiology text reports
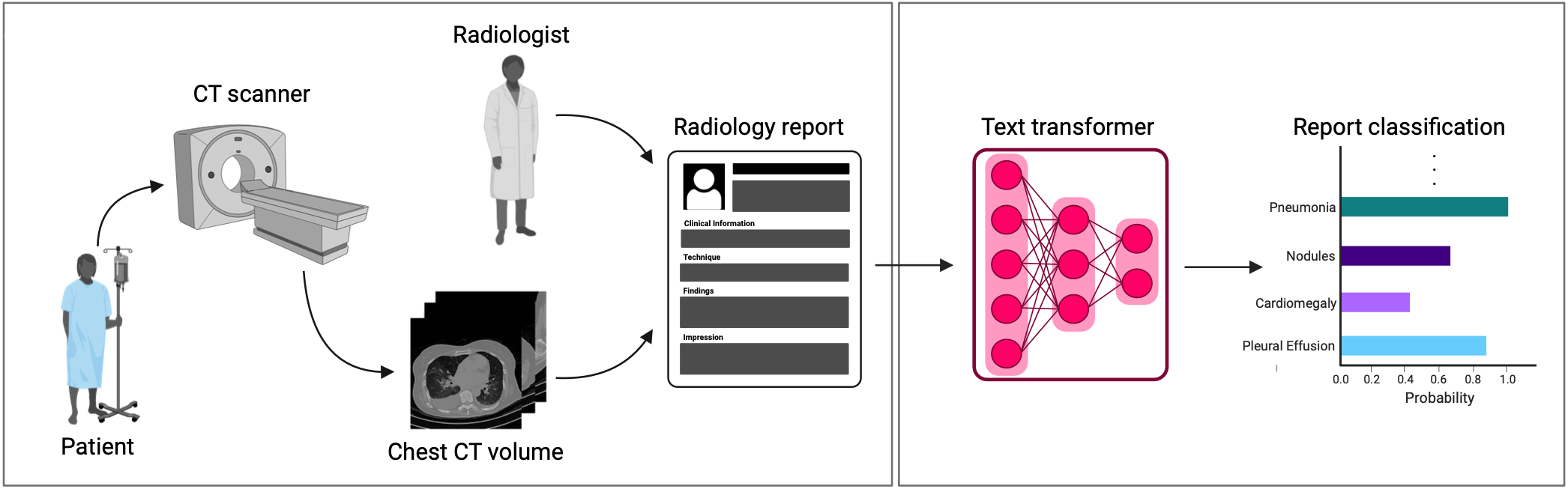
A major challenge in computational research in 3D medical imaging is the lack of comprehensive datasets. Addressing this issue, we present CT-RATE, the first 3D medical imaging dataset that pairs images with textual reports. CT-RATE consists of 25,692 non-contrast chest CT volumes, expanded to 50,188 through various reconstructions, from 21,304 unique patients, along with corresponding radiology text reports, multi-abnormality labels, and metadata.
We divided the cohort into two groups: 20,000 patients were allocated to the training set and 1,304 to the validation set. Our folders are structured as split_patientID_scanID_reconstructionID. For instance, "valid_53_a_1" indicates that this is a CT volume from the validation set, scan "a" from patient 53, and reconstruction 1 of scan "a". This naming convention applies to all files.
## CT-CLIP: CT-focused contrastive language-image pre-training framework

Leveraging CT-RATE, we developed CT-CLIP, a CT-focused contrastive language-image pre-training framework. As a versatile, self-supervised model, CT-CLIP is designed for broad application and does not require task-specific training. Remarkably, CT-CLIP outperforms state-of-the-art, fully supervised methods in multi-abnormality detection across all key metrics, thus eliminating the need for manual annotation. We also demonstrate its utility in case retrieval, whether using imagery or textual queries, thereby advancing knowledge dissemination.
Our complete codebase is openly available on [our official GitHub repository](https://github.com/ibrahimethemhamamci/CT-CLIP).
## CT-CHAT: Vision-language foundational chat model for 3D chest CT volumes

Leveraging [the VQA dataset](https://huggingface.co./datasets/ibrahimhamamci/CT-RATE/tree/main/dataset/vqa) derived from CT-RATE and pretrained 3D vision encoder from CT-CLIP, we developed CT-CHAT, a multimodal AI assistant designed to enhance the interpretation and diagnostic capabilities of 3D chest CT imaging. Building on the strong foundation of CT-CLIP, it integrates both visual and language processing to handle diverse tasks like visual question answering, report generation, and multiple-choice questions. Trained on over 2.7 million question-answer pairs from CT-RATE, it leverages 3D spatial information, making it superior to 2D-based models. CT-CHAT not only improves radiologist workflows by reducing interpretation time but also delivers highly accurate and clinically relevant responses, pushing the boundaries of 3D medical imaging tasks.
Our complete codebase is openly available on [our official GitHub repository](https://github.com/ibrahimethemhamamci/CT-CHAT).
## Citing Us
When using this dataset, please consider citing the following related papers:
```
1. @misc{hamamci2024foundation,
title={Developing Generalist Foundation Models from a Multimodal Dataset for 3D Computed Tomography},
author={Ibrahim Ethem Hamamci and Sezgin Er and Furkan Almas and Ayse Gulnihan Simsek and Sevval Nil Esirgun and Irem Dogan and Muhammed Furkan Dasdelen and Omer Faruk Durugol and Bastian Wittmann and Tamaz Amiranashvili and Enis Simsar and Mehmet Simsar and Emine Bensu Erdemir and Abdullah Alanbay and Anjany Sekuboyina and Berkan Lafci and Christian Bluethgen and Mehmet Kemal Ozdemir and Bjoern Menze},
year={2024},
eprint={2403.17834},
archivePrefix={arXiv},
primaryClass={cs.CV},
url={https://arxiv.org/abs/2403.17834},
}
(Accepted to ECCV 2024)
2. @misc{hamamci2024generatect,
title={GenerateCT: Text-Conditional Generation of 3D Chest CT Volumes},
author={Ibrahim Ethem Hamamci and Sezgin Er and Anjany Sekuboyina and Enis Simsar and Alperen Tezcan and Ayse Gulnihan Simsek and Sevval Nil Esirgun and Furkan Almas and Irem Dogan and Muhammed Furkan Dasdelen and Chinmay Prabhakar and Hadrien Reynaud and Sarthak Pati and Christian Bluethgen and Mehmet Kemal Ozdemir and Bjoern Menze},
year={2024},
eprint={2305.16037},
archivePrefix={arXiv},
primaryClass={cs.CV},
url={https://arxiv.org/abs/2305.16037},
}
(Accepted to MICCAI 2024)
3. @misc{hamamci2024ct2rep,
title={CT2Rep: Automated Radiology Report Generation for 3D Medical Imaging},
author={Ibrahim Ethem Hamamci and Sezgin Er and Bjoern Menze},
year={2024},
eprint={2403.06801},
archivePrefix={arXiv},
primaryClass={eess.IV},
url={https://arxiv.org/abs/2403.06801},
}
```
## Ethical Approval
For those who require ethical approval to apply for grants with this dataset, it can be accessed [here](./ethical_approval.PDF).
## License
We are committed to fostering innovation and collaboration in the research community. To this end, all elements of the CT-RATE dataset are released under a [Creative Commons Attribution (CC-BY-NC-SA) license](https://creativecommons.org/licenses/by-nc-sa/4.0/). This licensing framework ensures that our contributions can be freely used for non-commercial research purposes, while also encouraging contributions and modifications, provided that the original work is properly cited and any derivative works are shared under similar terms.


