Protein similarity and Matryoshka embeddings
Background
In 2022 UniProt added embeddings for each verified protein in their database. The embeddings represent the amino acid sequence which makes that protein, and can be used to measure similarity and make search applications.
Recently there's interest in the ML world to rethink embeddings to make vector databases faster and more efficient. After reading about Matryoshka embeddings, I came up with a project to train an adapted protein model. To summarize the concept, loss is organized for the earlier indexes in the embedding to hold more of the meaning. You can use a shorter embedding (instead of the full 1,024 dimensions used by UniProt) for approximate results. Projects in this space often do a first pass searching with shorter embeddings and then re-ranking based on the full embedding - Supabase blog post.
Embeddings data
I need to train the model on pairs of proteins with similarity scores derived from UniProt's current embeddings.
How to select the right pairs for training? I considered two factors:
- I need to pair within separate groups for training and testing. I decided to use the train/test/validation splits from the khairi/uniprot-swissprot dataset.
- Pairs should represent a mix of shorter and further differences (i.e. not just "these proteins don't look alike at all").
Let's look at 100,000 random pairs and get cosine distances. I was worried that the proteins would be clumped somewhere, but this is a cool distribution with a mean at 0.674.
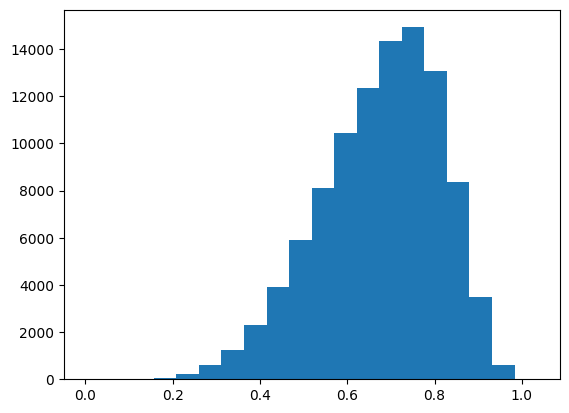
I figure for each protein I should pair it with two at random, then find ones with distance < 0.55 and > 0.8 (approximately top and bottom quintiles in distances of random pairs).
Dataset of protein pairs and distances: protein-pairs-uniprot-swissprot
CoLab Notebook: https://colab.research.google.com/drive/1rhoF0pAauHbiaNHdZkOI5O7A3mMcvS6G?usp=sharing
The agemagician/uniref50 dataset would be an alternative source for protein pairs, but it includes unreviewed/unverified proteins which UniProt did not pre-calculate embeddings for.
Training
UniProt made their embeddings with the Rostlab/prot_t5_xl_uniref50 model. For my base model, I use the same lab's smaller BERT model, prot_bert_bfd.
The tokenizer is based on IUPAC-IUB codes where each amino acid maps to a letter of the alphabet. NOTE: the text input must have spaces between each letter (e.g. "M S L E Q").
The good news is that this BERT base model is already pre-trained on amino acid sequences, and I just need a SentenceTransformers wrapper around the model to train with Matryoshka loss.
Once things were running smoothly on batches of 2 on a V100 GPU, I upped the batch size to 10 and ran it on an A100. I ran this for about 3.5 hours; it would take >20 hours for the full training set.
Embeddings model: https://huggingface.co./monsoon-nlp/protein-matryoshka-embeddings
CoLab Notebook: https://colab.research.google.com/drive/1uBk-jHOAPhIiUPPunfK7bMC8GnzpwmBy
Evaluation
During training, I used the SentenceTransformers EmbeddingSimilarityEvaluator to check that the full embeddings were working and improving on the validation set.
| steps | cosine_pearson | cosine_spearman |
|---|---|---|
| 3000 | 0.8598688660086558 | 0.8666855900999677 |
| 6000 | 0.8692703523988448 | 0.8615673651584274 |
| 9000 | 0.8779733537629968 | 0.8754158959780602 |
| 12000 | 0.8877422045031667 | 0.8881492475969834 |
| 15000 | 0.9027359688395733 | 0.899106724739699 |
| 18000 | 0.9046675789738002 | 0.9044183600191271 |
| 21000 | 0.9165801536390973 | 0.9061381997421003 |
| 24000 | 0.9128046401341833 | 0.9076748537082228 |
| 27000 | 0.918547416546341 | 0.9127677526055185 |
| 30000 | 0.9239429677657788 | 0.9187051589781693 |
We still need to verify that these embeddings are useful, even when shortened.
I took 2,000 random protein pairs from the test set and compared the cosine distances on the original (x) and new Matryoshka-ed model (y). There are a few things to ponder about this plot, but essential takeaway is that they correlate on the test set:
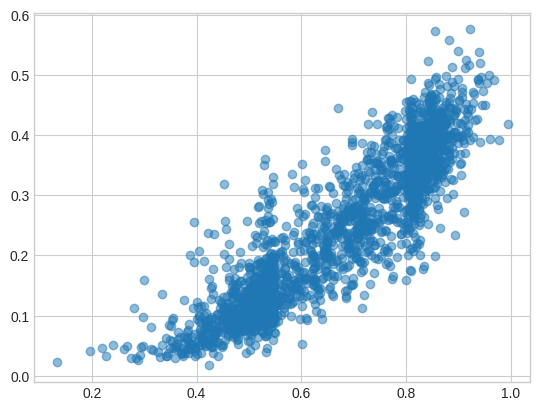
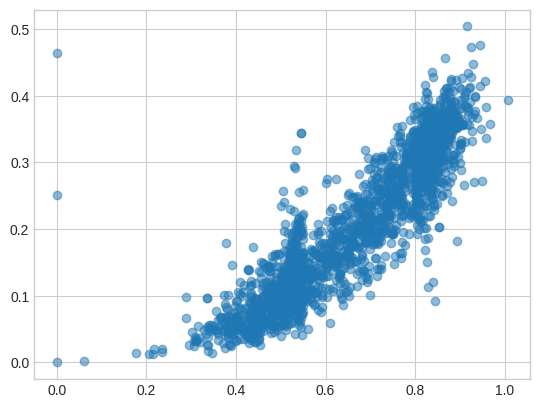
The plot for 128-dimension embeddings (different random pairs) looks quite similar!
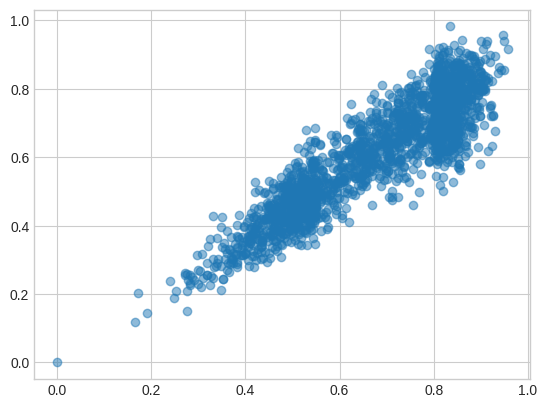
If I use the first 128 dimensions from the original model, they stick close to their full-size embeddings but there is noticeable drift:
CoLab Notebook: https://colab.research.google.com/drive/1hm4IIMXaLt_7QYRNvkiXl5BqmsHdC1Ue?usp=sharing
🦠🧬🤖🪆 Future Thoughts
Want to collaborate on bio LLMs? Get in touch!.
I want to do a project with plant DNA and possibly connect with genomics research for crops (potato, quinoa, wheat, rice, etc.).
Are the embeddings interesting enough to reach out to the UniProt team? Or to get resources to run the T5 model on the full dataset?
The Matryoshka paper tests the smaller embeddings on classification tasks, too. It looks like I could use MRL_Linear_Layer from this code to vary the embedding size. My current idea is to test it on tasks from TAPE.
While I was working on this, HuggingFace, Cohere, and pgvecto.rs had blog posts about quantized embeddings? Perhaps this would be useful, too.