daliprf
commited on
Commit
·
3dbf98e
1
Parent(s):
f7f2fc9
init
Browse files- .gitattributes +3 -0
- Data_custom_generator.py +56 -0
- README.md +150 -3
- cnn_model.py +94 -0
- configuration.py +77 -0
- custom_Losses.py +63 -0
- image_utility.py +585 -0
- img_printer.py +81 -0
- main.py +45 -0
- models/students/student_Net_300w.h5 +3 -0
- models/students/student_Net_COFW.h5 +3 -0
- models/students/student_Net_WFLW.h5 +3 -0
- pca_utility.py +286 -0
- requirements.txt +23 -0
- samples/KDLoss-1.jpg +3 -0
- samples/KD_300W_samples-1.jpg +3 -0
- samples/KD_L2_L1-1.jpg +3 -0
- samples/KD_WFLW_samples-1.jpg +3 -0
- samples/KD_cofw_samples-1.jpg +3 -0
- samples/general_framework-1.jpg +3 -0
- samples/loss_weight-1.jpg +3 -0
- samples/main_loss-1.jpg +3 -0
- samples/teacher_arch-1.jpg +3 -0
- student_train.py +292 -0
- teacher_trainer.py +215 -0
- test.py +40 -0
.gitattributes
CHANGED
|
@@ -29,3 +29,6 @@ saved_model/**/* filter=lfs diff=lfs merge=lfs -text
|
|
| 29 |
*.zip filter=lfs diff=lfs merge=lfs -text
|
| 30 |
*.zstandard filter=lfs diff=lfs merge=lfs -text
|
| 31 |
*tfevents* filter=lfs diff=lfs merge=lfs -text
|
|
|
|
|
|
|
|
|
|
|
|
| 29 |
*.zip filter=lfs diff=lfs merge=lfs -text
|
| 30 |
*.zstandard filter=lfs diff=lfs merge=lfs -text
|
| 31 |
*tfevents* filter=lfs diff=lfs merge=lfs -text
|
| 32 |
+
*.pdf filter=lfs diff=lfs merge=lfs -text
|
| 33 |
+
*.png filter=lfs diff=lfs merge=lfs -text
|
| 34 |
+
*.jpg filter=lfs diff=lfs merge=lfs -text
|
Data_custom_generator.py
ADDED
|
@@ -0,0 +1,56 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
from skimage.io import imread
|
| 2 |
+
from skimage.transform import resize
|
| 3 |
+
import numpy as np
|
| 4 |
+
import tensorflow as tf
|
| 5 |
+
import keras
|
| 6 |
+
from skimage.transform import resize
|
| 7 |
+
from tf_record_utility import TFRecordUtility
|
| 8 |
+
from configuration import DatasetName, DatasetType, \
|
| 9 |
+
AffectnetConf, D300wConf, W300Conf, InputDataSize, LearningConfig
|
| 10 |
+
from numpy import save, load, asarray
|
| 11 |
+
|
| 12 |
+
|
| 13 |
+
class CustomHeatmapGenerator(keras.utils.Sequence):
|
| 14 |
+
|
| 15 |
+
def __init__(self, is_single, image_filenames, label_filenames, batch_size, n_outputs, accuracy=100):
|
| 16 |
+
self.image_filenames = image_filenames
|
| 17 |
+
self.label_filenames = label_filenames
|
| 18 |
+
self.batch_size = batch_size
|
| 19 |
+
self.n_outputs = n_outputs
|
| 20 |
+
self.is_single = is_single
|
| 21 |
+
self.accuracy = accuracy
|
| 22 |
+
|
| 23 |
+
def __len__(self):
|
| 24 |
+
_len = np.ceil(len(self.image_filenames) // float(self.batch_size))
|
| 25 |
+
return int(_len)
|
| 26 |
+
|
| 27 |
+
def __getitem__(self, idx):
|
| 28 |
+
img_path = D300wConf.train_images_dir
|
| 29 |
+
tr_path_85 = D300wConf.train_hm_dir_85
|
| 30 |
+
tr_path_90 = D300wConf.train_hm_dir_90
|
| 31 |
+
tr_path_97 = D300wConf.train_hm_dir_97
|
| 32 |
+
tr_path = D300wConf.train_hm_dir
|
| 33 |
+
|
| 34 |
+
batch_x = self.image_filenames[idx * self.batch_size:(idx + 1) * self.batch_size]
|
| 35 |
+
batch_y = self.label_filenames[idx * self.batch_size:(idx + 1) * self.batch_size]
|
| 36 |
+
|
| 37 |
+
img_batch = np.array([imread(img_path + file_name) for file_name in batch_x])
|
| 38 |
+
|
| 39 |
+
if self.is_single:
|
| 40 |
+
if self.accuracy == 85:
|
| 41 |
+
lbl_batch = np.array([load(tr_path_85 + file_name) for file_name in batch_y])
|
| 42 |
+
elif self.accuracy == 90:
|
| 43 |
+
lbl_batch = np.array([load(tr_path_90 + file_name) for file_name in batch_y])
|
| 44 |
+
elif self.accuracy == 97:
|
| 45 |
+
lbl_batch = np.array([load(tr_path_97 + file_name) for file_name in batch_y])
|
| 46 |
+
else:
|
| 47 |
+
lbl_batch = np.array([load(tr_path + file_name) for file_name in batch_y])
|
| 48 |
+
|
| 49 |
+
lbl_out_array = lbl_batch
|
| 50 |
+
else:
|
| 51 |
+
lbl_batch_85 = np.array([load(tr_path_85 + file_name) for file_name in batch_y])
|
| 52 |
+
lbl_batch_90 = np.array([load(tr_path_90 + file_name) for file_name in batch_y])
|
| 53 |
+
lbl_batch_97 = np.array([load(tr_path_97 + file_name) for file_name in batch_y])
|
| 54 |
+
lbl_out_array = [lbl_batch_85, lbl_batch_90, lbl_batch_97]
|
| 55 |
+
|
| 56 |
+
return img_batch, lbl_out_array
|
README.md
CHANGED
|
@@ -1,3 +1,150 @@
|
|
| 1 |
-
|
| 2 |
-
|
| 3 |
-
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
|
| 2 |
+
[](https://paperswithcode.com/sota/face-alignment-on-cofw?p=facial-landmark-points-detection-using)
|
| 3 |
+
|
| 4 |
+
#
|
| 5 |
+
Facial Landmark Points Detection Using Knowledge Distillation-Based Neural Networks
|
| 6 |
+
|
| 7 |
+
|
| 8 |
+
|
| 9 |
+
#### Link to the paper:
|
| 10 |
+
Google Scholar:
|
| 11 |
+
https://scholar.google.com/citations?view_op=view_citation&hl=en&user=96lS6HIAAAAJ&citation_for_view=96lS6HIAAAAJ:zYLM7Y9cAGgC
|
| 12 |
+
|
| 13 |
+
Elsevier:
|
| 14 |
+
https://www.sciencedirect.com/science/article/pii/S1077314221001582
|
| 15 |
+
|
| 16 |
+
Arxiv:
|
| 17 |
+
https://arxiv.org/abs/2111.07047
|
| 18 |
+
|
| 19 |
+
#### Link to the paperswithcode.com:
|
| 20 |
+
https://paperswithcode.com/paper/facial-landmark-points-detection-using
|
| 21 |
+
|
| 22 |
+
```diff
|
| 23 |
+
@@plaese STAR the repo if you like it.@@
|
| 24 |
+
```
|
| 25 |
+
|
| 26 |
+
```
|
| 27 |
+
Please cite this work as:
|
| 28 |
+
|
| 29 |
+
@article{fard2022facial,
|
| 30 |
+
title={Facial landmark points detection using knowledge distillation-based neural networks},
|
| 31 |
+
author={Fard, Ali Pourramezan and Mahoor, Mohammad H},
|
| 32 |
+
journal={Computer Vision and Image Understanding},
|
| 33 |
+
volume={215},
|
| 34 |
+
pages={103316},
|
| 35 |
+
year={2022},
|
| 36 |
+
publisher={Elsevier}
|
| 37 |
+
}
|
| 38 |
+
|
| 39 |
+
```
|
| 40 |
+
|
| 41 |
+
## Introduction
|
| 42 |
+
Facial landmark detection is a vital step for numerous facial image analysis applications. Although some deep learning-based methods have achieved good performances in this task, they are often not suitable for running on mobile devices. Such methods rely on networks with many parameters, which makes the training and inference time-consuming. Training lightweight neural networks such as MobileNets are often challenging, and the models might have low accuracy. Inspired by knowledge distillation (KD), this paper presents a novel loss function to train a lightweight Student network (e.g., MobileNetV2) for facial landmark detection. We use two Teacher networks, a Tolerant-Teacher and a Tough-Teacher in conjunction with the Student network. The Tolerant-Teacher is trained using Soft-landmarks created by active shape models, while the Tough-Teacher is trained using the ground truth (aka Hard-landmarks) landmark points. To utilize the facial landmark points predicted by the Teacher networks, we define an Assistive Loss (ALoss) for each Teacher network. Moreover, we define a loss function called KD-Loss that utilizes the facial landmark points predicted by the two pre-trained Teacher networks (EfficientNet-b3) to guide the lightweight Student network towards predicting the Hard-landmarks. Our experimental results on three challenging facial datasets show that the proposed architecture will result in a better-trained Student network that can extract facial landmark points with high accuracy.
|
| 43 |
+
|
| 44 |
+
|
| 45 |
+
##Architecture
|
| 46 |
+
|
| 47 |
+
We train the Tough-Teacher, and the Tolerant-Teacher networks independently using the Hard-landmarks and the Soft-landmarks respectively utilizing the L2 loss:
|
| 48 |
+
|
| 49 |
+
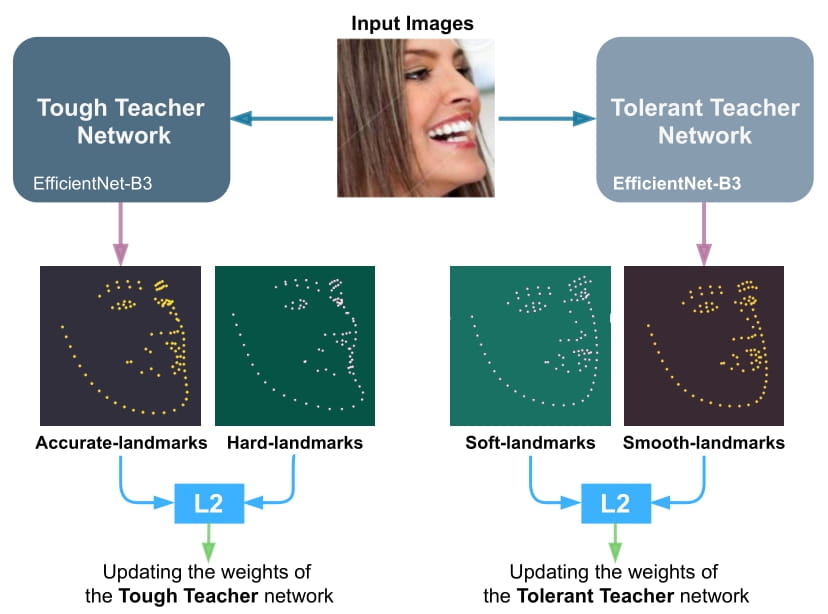
|
| 50 |
+
|
| 51 |
+
|
| 52 |
+
Proposed KD-based architecture for training the Student network. KDLoss uses the knowledge of the previously trained Teacher networks by utilizing the assistive loss functions ALossT ou and ALossT ol, to improve the performance the face alignment task:
|
| 53 |
+
|
| 54 |
+
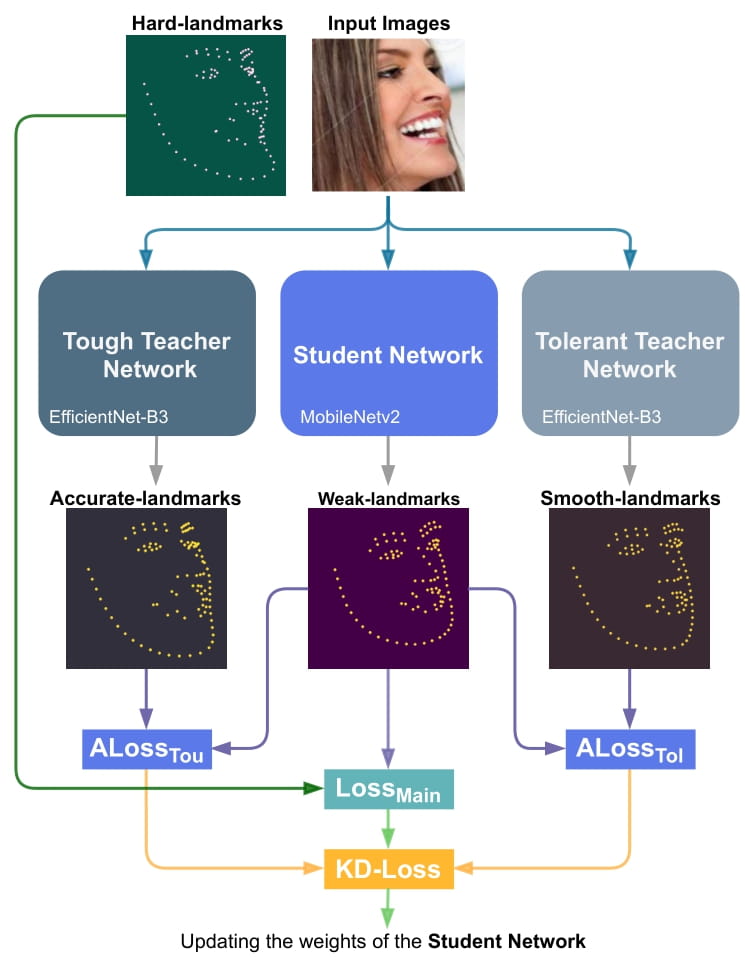
|
| 55 |
+
|
| 56 |
+
|
| 57 |
+
## Evaluation
|
| 58 |
+
|
| 59 |
+
Following are some samples in order to show the visual performance of KD-Loss on 300W, COFW and WFLW datasets:
|
| 60 |
+
|
| 61 |
+
300W:
|
| 62 |
+

|
| 63 |
+
|
| 64 |
+
COFW:
|
| 65 |
+

|
| 66 |
+
|
| 67 |
+
WFLW:
|
| 68 |
+

|
| 69 |
+
|
| 70 |
+
----------------------------------------------------------------------------------------------------------------------------------
|
| 71 |
+
## Installing the requirements
|
| 72 |
+
In order to run the code you need to install python >= 3.5.
|
| 73 |
+
The requirements and the libraries needed to run the code can be installed using the following command:
|
| 74 |
+
|
| 75 |
+
```
|
| 76 |
+
pip install -r requirements.txt
|
| 77 |
+
```
|
| 78 |
+
|
| 79 |
+
|
| 80 |
+
## Using the pre-trained models
|
| 81 |
+
You can test and use the preetrained models using the following codes which are available in the test.py:
|
| 82 |
+
The pretrained student model are also located in "models/students".
|
| 83 |
+
|
| 84 |
+
```
|
| 85 |
+
cnn = CNNModel()
|
| 86 |
+
model = cnn.get_model(arch=arch, input_tensor=None, output_len=self.output_len)
|
| 87 |
+
|
| 88 |
+
model.load_weights(weight_fname)
|
| 89 |
+
|
| 90 |
+
img = None # load a cropped image
|
| 91 |
+
|
| 92 |
+
image_utility = ImageUtility()
|
| 93 |
+
pose_predicted = []
|
| 94 |
+
image = np.expand_dims(img, axis=0)
|
| 95 |
+
|
| 96 |
+
pose_predicted = model.predict(image)[1][0]
|
| 97 |
+
```
|
| 98 |
+
|
| 99 |
+
|
| 100 |
+
## Training Network from scratch
|
| 101 |
+
|
| 102 |
+
|
| 103 |
+
### Preparing Data
|
| 104 |
+
Data needs to be normalized and saved in npy format.
|
| 105 |
+
|
| 106 |
+
### Training
|
| 107 |
+
|
| 108 |
+
### Training Teacher Networks:
|
| 109 |
+
|
| 110 |
+
The training implementation is located in teacher_trainer.py class. You can use the following code to start the training for the teacher networks:
|
| 111 |
+
|
| 112 |
+
```
|
| 113 |
+
'''train Teacher Networks'''
|
| 114 |
+
trainer = TeacherTrainer(dataset_name=DatasetName.w300)
|
| 115 |
+
trainer.train(arch='efficientNet',weight_path=None)
|
| 116 |
+
```
|
| 117 |
+
|
| 118 |
+
### Training Student Networks:
|
| 119 |
+
After Training the teacher networks, you can use the trained teachers to train the student network. The implemetation of training of the student network is provided in teacher_trainer.py . You can use the following code to start the training for the student networks:
|
| 120 |
+
|
| 121 |
+
```
|
| 122 |
+
st_trainer = StudentTrainer(dataset_name=DatasetName.w300, use_augmneted=True)
|
| 123 |
+
st_trainer.train(arch_student='mobileNetV2', weight_path_student=None,
|
| 124 |
+
loss_weight_student=2.0,
|
| 125 |
+
arch_tough_teacher='efficientNet', weight_path_tough_teacher='./models/teachers/ds_300w_ef_tou.h5',
|
| 126 |
+
loss_weight_tough_teacher=1,
|
| 127 |
+
arch_tol_teacher='efficientNet', weight_path_tol_teacher='./models/teachers/ds_300w_ef_tol.h5',
|
| 128 |
+
loss_weight_tol_teacher=1)
|
| 129 |
+
```
|
| 130 |
+
|
| 131 |
+
|
| 132 |
+
|
| 133 |
+
```
|
| 134 |
+
Please cite this work as:
|
| 135 |
+
|
| 136 |
+
@article{fard2022facial,
|
| 137 |
+
title={Facial landmark points detection using knowledge distillation-based neural networks},
|
| 138 |
+
author={Fard, Ali Pourramezan and Mahoor, Mohammad H},
|
| 139 |
+
journal={Computer Vision and Image Understanding},
|
| 140 |
+
volume={215},
|
| 141 |
+
pages={103316},
|
| 142 |
+
year={2022},
|
| 143 |
+
publisher={Elsevier}
|
| 144 |
+
}
|
| 145 |
+
|
| 146 |
+
```
|
| 147 |
+
|
| 148 |
+
```diff
|
| 149 |
+
@@plaese STAR the repo if you like it.@@
|
| 150 |
+
```
|
cnn_model.py
ADDED
|
@@ -0,0 +1,94 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
from configuration import DatasetName, DatasetType, \
|
| 2 |
+
D300wConf, InputDataSize, LearningConfig
|
| 3 |
+
|
| 4 |
+
import tensorflow as tf
|
| 5 |
+
# tf.compat.v1.disable_eager_execution()
|
| 6 |
+
|
| 7 |
+
from tensorflow import keras
|
| 8 |
+
from skimage.transform import resize
|
| 9 |
+
from keras.regularizers import l2
|
| 10 |
+
|
| 11 |
+
# tf.logging.set_verbosity(tf.logging.ERROR)
|
| 12 |
+
from tensorflow.keras.models import Model
|
| 13 |
+
from tensorflow.keras.applications import mobilenet_v2, mobilenet, resnet50, densenet
|
| 14 |
+
|
| 15 |
+
from tensorflow.keras.layers import Dense, MaxPooling2D, Conv2D, Flatten, \
|
| 16 |
+
BatchNormalization, Activation, GlobalAveragePooling2D, DepthwiseConv2D, Dropout, ReLU, Concatenate, Input, Conv2DTranspose
|
| 17 |
+
|
| 18 |
+
from keras.callbacks import ModelCheckpoint
|
| 19 |
+
from keras import backend as K
|
| 20 |
+
|
| 21 |
+
from keras.optimizers import Adam
|
| 22 |
+
import numpy as np
|
| 23 |
+
import matplotlib.pyplot as plt
|
| 24 |
+
import math
|
| 25 |
+
from keras.callbacks import CSVLogger
|
| 26 |
+
from datetime import datetime
|
| 27 |
+
|
| 28 |
+
import cv2
|
| 29 |
+
import os.path
|
| 30 |
+
from keras.utils.vis_utils import plot_model
|
| 31 |
+
from scipy.spatial import distance
|
| 32 |
+
import scipy.io as sio
|
| 33 |
+
|
| 34 |
+
import efficientnet.tfkeras as efn
|
| 35 |
+
|
| 36 |
+
|
| 37 |
+
class CNNModel:
|
| 38 |
+
def get_model(self, arch, input_tensor, output_len,
|
| 39 |
+
inp_shape=[InputDataSize.image_input_size, InputDataSize.image_input_size, 3],
|
| 40 |
+
weight_path=None):
|
| 41 |
+
if arch == 'efficientNet':
|
| 42 |
+
model = self.create_efficientNet(inp_shape=inp_shape, input_tensor=input_tensor, output_len=output_len)
|
| 43 |
+
elif arch == 'mobileNetV2':
|
| 44 |
+
model = self.create_MobileNet(inp_shape=inp_shape, inp_tensor=input_tensor)
|
| 45 |
+
return model
|
| 46 |
+
|
| 47 |
+
def create_MobileNet(self, inp_shape, inp_tensor):
|
| 48 |
+
|
| 49 |
+
mobilenet_model = mobilenet_v2.MobileNetV2(input_shape=inp_shape,
|
| 50 |
+
alpha=1.0,
|
| 51 |
+
include_top=True,
|
| 52 |
+
weights=None,
|
| 53 |
+
input_tensor=inp_tensor,
|
| 54 |
+
pooling=None)
|
| 55 |
+
|
| 56 |
+
inp = mobilenet_model.input
|
| 57 |
+
out_landmarks = mobilenet_model.get_layer('O_L').output
|
| 58 |
+
revised_model = Model(inp, [out_landmarks])
|
| 59 |
+
model_json = revised_model.to_json()
|
| 60 |
+
with open("mobileNet_v2_stu.json", "w") as json_file:
|
| 61 |
+
json_file.write(model_json)
|
| 62 |
+
return revised_model
|
| 63 |
+
|
| 64 |
+
def create_efficientNet(self, inp_shape, input_tensor, output_len, is_teacher=True):
|
| 65 |
+
if is_teacher: # for teacher we use a heavier network
|
| 66 |
+
eff_net = efn.EfficientNetB3(include_top=True,
|
| 67 |
+
weights=None,
|
| 68 |
+
input_tensor=None,
|
| 69 |
+
input_shape=[InputDataSize.image_input_size, InputDataSize.image_input_size,
|
| 70 |
+
3],
|
| 71 |
+
pooling=None,
|
| 72 |
+
classes=output_len)
|
| 73 |
+
else: # for student we use the small network
|
| 74 |
+
eff_net = efn.EfficientNetB0(include_top=True,
|
| 75 |
+
weights=None,
|
| 76 |
+
input_tensor=None,
|
| 77 |
+
input_shape=inp_shape,
|
| 78 |
+
pooling=None,
|
| 79 |
+
classes=output_len) # or weights='noisy-student'
|
| 80 |
+
|
| 81 |
+
eff_net.layers.pop()
|
| 82 |
+
inp = eff_net.input
|
| 83 |
+
|
| 84 |
+
x = eff_net.get_layer('top_activation').output
|
| 85 |
+
x = GlobalAveragePooling2D()(x)
|
| 86 |
+
x = keras.layers.Dropout(rate=0.5)(x)
|
| 87 |
+
output = Dense(output_len, activation='linear', name='out')(x)
|
| 88 |
+
|
| 89 |
+
eff_net = Model(inp, output)
|
| 90 |
+
|
| 91 |
+
eff_net.summary()
|
| 92 |
+
|
| 93 |
+
return eff_net
|
| 94 |
+
|
configuration.py
ADDED
|
@@ -0,0 +1,77 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
class DatasetName:
|
| 2 |
+
w300 = 'w300'
|
| 3 |
+
cofw = 'cofw'
|
| 4 |
+
wflw = 'wflw'
|
| 5 |
+
|
| 6 |
+
class DatasetType:
|
| 7 |
+
data_type_train = 0
|
| 8 |
+
data_type_validation = 1
|
| 9 |
+
data_type_test = 2
|
| 10 |
+
|
| 11 |
+
class LearningConfig:
|
| 12 |
+
batch_size = 70
|
| 13 |
+
epochs = 150
|
| 14 |
+
|
| 15 |
+
class InputDataSize:
|
| 16 |
+
image_input_size = 224
|
| 17 |
+
|
| 18 |
+
class WflwConf:
|
| 19 |
+
Wflw_prefix_path = './data/wflw/' # --> local
|
| 20 |
+
|
| 21 |
+
''' augmented version'''
|
| 22 |
+
augmented_train_pose = Wflw_prefix_path + 'training_set/augmented/pose/'
|
| 23 |
+
augmented_train_annotation = Wflw_prefix_path + 'training_set/augmented/annotations/'
|
| 24 |
+
augmented_train_atr = Wflw_prefix_path + 'training_set/augmented/atrs/'
|
| 25 |
+
augmented_train_image = Wflw_prefix_path + 'training_set/augmented/images/'
|
| 26 |
+
augmented_train_tf_path = Wflw_prefix_path + 'training_set/augmented/tf/'
|
| 27 |
+
''' original version'''
|
| 28 |
+
no_aug_train_annotation = Wflw_prefix_path + 'training_set/no_aug/annotations/'
|
| 29 |
+
no_aug_train_atr = Wflw_prefix_path + 'training_set/no_aug/atrs/'
|
| 30 |
+
no_aug_train_pose = Wflw_prefix_path + 'training_set/no_aug/pose/'
|
| 31 |
+
no_aug_train_image = Wflw_prefix_path + 'training_set/no_aug/images/'
|
| 32 |
+
no_aug_train_tf_path = Wflw_prefix_path + 'training_set/no_aug/tf/'
|
| 33 |
+
|
| 34 |
+
orig_number_of_training = 7500
|
| 35 |
+
orig_number_of_test = 2500
|
| 36 |
+
|
| 37 |
+
augmentation_factor = 4 # create . image from 1
|
| 38 |
+
augmentation_factor_rotate = 15 # create . image from 1
|
| 39 |
+
num_of_landmarks = 98
|
| 40 |
+
|
| 41 |
+
class CofwConf:
|
| 42 |
+
Cofw_prefix_path = './data/cofw/' # --> local
|
| 43 |
+
|
| 44 |
+
augmented_train_pose = Cofw_prefix_path + 'training_set/augmented/pose/'
|
| 45 |
+
augmented_train_annotation = Cofw_prefix_path + 'training_set/augmented/annotations/'
|
| 46 |
+
augmented_train_image = Cofw_prefix_path + 'training_set/augmented/images/'
|
| 47 |
+
augmented_train_tf_path = Cofw_prefix_path + 'training_set/augmented/tf/'
|
| 48 |
+
|
| 49 |
+
orig_number_of_training = 1345
|
| 50 |
+
orig_number_of_test = 507
|
| 51 |
+
|
| 52 |
+
augmentation_factor = 10
|
| 53 |
+
num_of_landmarks = 29
|
| 54 |
+
|
| 55 |
+
|
| 56 |
+
class D300wConf:
|
| 57 |
+
w300w_prefix_path = './data/300W/' # --> local
|
| 58 |
+
|
| 59 |
+
orig_300W_train = w300w_prefix_path + 'orig_300W_train/'
|
| 60 |
+
augmented_train_pose = w300w_prefix_path + 'training_set/augmented/pose/'
|
| 61 |
+
augmented_train_annotation = w300w_prefix_path + 'training_set/augmented/annotations/'
|
| 62 |
+
augmented_train_image = w300w_prefix_path + 'training_set/augmented/images/'
|
| 63 |
+
augmented_train_tf_path = w300w_prefix_path + 'training_set/augmented/tf/'
|
| 64 |
+
|
| 65 |
+
no_aug_train_annotation = w300w_prefix_path + 'training_set/no_aug/annotations/'
|
| 66 |
+
no_aug_train_pose = w300w_prefix_path + 'training_set/no_aug/pose/'
|
| 67 |
+
no_aug_train_image = w300w_prefix_path + 'training_set/no_aug/images/'
|
| 68 |
+
no_aug_train_tf_path = w300w_prefix_path + 'training_set/no_aug/tf/'
|
| 69 |
+
|
| 70 |
+
orig_number_of_training = 3148
|
| 71 |
+
orig_number_of_test_full = 689
|
| 72 |
+
orig_number_of_test_common = 554
|
| 73 |
+
orig_number_of_test_challenging = 135
|
| 74 |
+
|
| 75 |
+
augmentation_factor = 4 # create . image from 1
|
| 76 |
+
num_of_landmarks = 68
|
| 77 |
+
|
custom_Losses.py
ADDED
|
@@ -0,0 +1,63 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import math
|
| 2 |
+
|
| 3 |
+
import numpy as np
|
| 4 |
+
import tensorflow as tf
|
| 5 |
+
|
| 6 |
+
# tf.compat.v1.disable_eager_execution()
|
| 7 |
+
# tf.compat.v1.enable_eager_execution()
|
| 8 |
+
|
| 9 |
+
from PIL import Image
|
| 10 |
+
from tensorflow.keras import backend as K
|
| 11 |
+
from scipy.spatial import distance
|
| 12 |
+
|
| 13 |
+
from cnn_model import CNNModel
|
| 14 |
+
from configuration import DatasetName, D300wConf, LearningConfig
|
| 15 |
+
from image_utility import ImageUtility
|
| 16 |
+
from pca_utility import PCAUtility
|
| 17 |
+
|
| 18 |
+
print(tf.__version__)
|
| 19 |
+
|
| 20 |
+
|
| 21 |
+
class Custom_losses:
|
| 22 |
+
|
| 23 |
+
def MSE(self, x_pr, x_gt):
|
| 24 |
+
return tf.losses.mean_squared_error(x_pr, x_gt)
|
| 25 |
+
|
| 26 |
+
|
| 27 |
+
def kd_loss(self, x_pr, x_gt, x_tough, x_tol, alpha_tough, alpha_mi_tough, alpha_tol, alpha_mi_tol,
|
| 28 |
+
main_loss_weight, tough_loss_weight, tol_loss_weight):
|
| 29 |
+
"""km"""
|
| 30 |
+
'''los KD'''
|
| 31 |
+
# we revise teachers for reflection:
|
| 32 |
+
x_tough = x_gt + tf.sign(x_pr - x_gt) * tf.abs(x_tough - x_gt)
|
| 33 |
+
b_tough = x_gt + tf.sign(x_pr - x_gt) * tf.abs(x_tough - x_gt) * 0.15
|
| 34 |
+
x_tol = x_gt + tf.sign(x_pr - x_gt) * tf.abs(x_tol - x_gt)
|
| 35 |
+
b_tol = x_gt + tf.sign(x_pr - x_gt) * tf.abs(x_tol - x_gt) * 0.15
|
| 36 |
+
# Region A: from T -> +inf
|
| 37 |
+
tou_pos_map = tf.where(tf.sign(x_pr - x_tough) * tf.sign(x_tough - x_gt) > 0, alpha_tough, 0.0)
|
| 38 |
+
tou_neg_map = tf.where(tf.sign(x_tough - x_pr) * tf.sign(x_pr - b_tough) >= 0, alpha_mi_tough, 0.0)
|
| 39 |
+
# tou_red_map = tf.where(tf.sign(tf.abs(b_tough) - tf.abs(x_pr))*tf.sign(tf.abs(x_pr) - tf.abs(x_gt)) > 0, 0.1, 0.0)
|
| 40 |
+
tou_map = tou_pos_map + tou_neg_map # + tou_red_map
|
| 41 |
+
|
| 42 |
+
tol_pos_map = tf.where(tf.sign(x_pr - x_tol) * tf.sign(x_tol - x_gt) > 0, alpha_tol, 0.0)
|
| 43 |
+
tol_neg_map = tf.where(tf.sign(x_tol - x_pr) * tf.sign(x_pr - b_tol) >= 0, alpha_mi_tol, 0.0)
|
| 44 |
+
# tol_red_map = tf.where(tf.sign(tf.abs(b_tol) - tf.abs(x_pr))*tf.sign(tf.abs(x_pr) - tf.abs(x_gt)) > 0, 0.1, 0.0)
|
| 45 |
+
tol_map = tol_pos_map + tol_neg_map # + tou_red_map
|
| 46 |
+
|
| 47 |
+
'''calculate dif map for linear and non-linear part'''
|
| 48 |
+
low_diff_main_map = tf.where(tf.abs(x_gt - x_pr) <= tf.abs(x_gt - x_tol), 1.0, 0.0)
|
| 49 |
+
high_diff_main_map = tf.where(tf.abs(x_gt - x_pr) > tf.abs(x_gt - x_tol), 1.0, 0.0)
|
| 50 |
+
|
| 51 |
+
'''calculate loss'''
|
| 52 |
+
loss_main_high_dif = tf.reduce_mean(
|
| 53 |
+
high_diff_main_map * (tf.square(x_gt - x_pr) + (3 * tf.abs(x_gt - x_tol)) - tf.square(x_gt - x_tol)))
|
| 54 |
+
loss_main_low_dif = tf.reduce_mean(low_diff_main_map * (3 * tf.abs(x_gt - x_pr)))
|
| 55 |
+
loss_main = main_loss_weight * (loss_main_high_dif + loss_main_low_dif)
|
| 56 |
+
|
| 57 |
+
loss_tough_assist = tough_loss_weight * tf.reduce_mean(tou_map * tf.abs(x_tough - x_pr))
|
| 58 |
+
loss_tol_assist = tol_loss_weight * tf.reduce_mean(tol_map * tf.abs(x_tol - x_pr))
|
| 59 |
+
|
| 60 |
+
'''dif loss:'''
|
| 61 |
+
loss_total = loss_main + loss_tough_assist + loss_tol_assist
|
| 62 |
+
|
| 63 |
+
return loss_total, loss_main, loss_tough_assist, loss_tol_assist
|
image_utility.py
ADDED
|
@@ -0,0 +1,585 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import random
|
| 2 |
+
import numpy as np
|
| 3 |
+
|
| 4 |
+
import matplotlib
|
| 5 |
+
matplotlib.use('agg')
|
| 6 |
+
import matplotlib.pyplot as plt
|
| 7 |
+
|
| 8 |
+
import math
|
| 9 |
+
from skimage.transform import warp, AffineTransform
|
| 10 |
+
import cv2
|
| 11 |
+
from scipy import misc
|
| 12 |
+
from skimage.transform import rotate
|
| 13 |
+
from PIL import Image
|
| 14 |
+
from PIL import ImageOps
|
| 15 |
+
from skimage.transform import resize
|
| 16 |
+
from skimage import transform
|
| 17 |
+
from skimage.transform import SimilarityTransform, AffineTransform
|
| 18 |
+
import random
|
| 19 |
+
|
| 20 |
+
class ImageUtility:
|
| 21 |
+
|
| 22 |
+
def random_rotate(self, _image, _label, file_name, fn):
|
| 23 |
+
|
| 24 |
+
xy_points, x_points, y_points = self.create_landmarks(landmarks=_label,
|
| 25 |
+
scale_factor_x=1, scale_factor_y=1)
|
| 26 |
+
_image, _label = self.cropImg_2time(_image, x_points, y_points)
|
| 27 |
+
|
| 28 |
+
scale = (np.random.uniform(0.7, 1.3), np.random.uniform(0.7, 1.3))
|
| 29 |
+
# scale = (1, 1)
|
| 30 |
+
|
| 31 |
+
rot = np.random.uniform(-1 * 0.55, 0.55)
|
| 32 |
+
translation = (0, 0)
|
| 33 |
+
shear = 0
|
| 34 |
+
|
| 35 |
+
tform = AffineTransform(
|
| 36 |
+
scale=scale, # ,
|
| 37 |
+
rotation=rot,
|
| 38 |
+
translation=translation,
|
| 39 |
+
shear=np.deg2rad(shear)
|
| 40 |
+
)
|
| 41 |
+
|
| 42 |
+
output_img = transform.warp(_image, tform.inverse, mode='symmetric')
|
| 43 |
+
|
| 44 |
+
sx, sy = scale
|
| 45 |
+
t_matrix = np.array([
|
| 46 |
+
[sx * math.cos(rot), -sy * math.sin(rot + shear), 0],
|
| 47 |
+
[sx * math.sin(rot), sy * math.cos(rot + shear), 0],
|
| 48 |
+
[0, 0, 1]
|
| 49 |
+
])
|
| 50 |
+
landmark_arr_xy, landmark_arr_x, landmark_arr_y = self.create_landmarks(_label, 1, 1)
|
| 51 |
+
label = np.array(landmark_arr_x + landmark_arr_y).reshape([2, 68])
|
| 52 |
+
marging = np.ones([1, 68])
|
| 53 |
+
label = np.concatenate((label, marging), axis=0)
|
| 54 |
+
|
| 55 |
+
label_t = np.dot(t_matrix, label)
|
| 56 |
+
lbl_flat = np.delete(label_t, 2, axis=0).reshape([136])
|
| 57 |
+
|
| 58 |
+
t_label = self.__reorder(lbl_flat)
|
| 59 |
+
|
| 60 |
+
'''crop data: we add a small margin to the images'''
|
| 61 |
+
xy_points, x_points, y_points = self.create_landmarks(landmarks=t_label,
|
| 62 |
+
scale_factor_x=1, scale_factor_y=1)
|
| 63 |
+
img_arr, points_arr = self.cropImg(output_img, x_points, y_points, no_padding=False)
|
| 64 |
+
# img_arr = output_img
|
| 65 |
+
# points_arr = t_label
|
| 66 |
+
'''resize image to 224*224'''
|
| 67 |
+
resized_img = resize(img_arr,
|
| 68 |
+
(224, 224, 3),
|
| 69 |
+
anti_aliasing=True)
|
| 70 |
+
dims = img_arr.shape
|
| 71 |
+
height = dims[0]
|
| 72 |
+
width = dims[1]
|
| 73 |
+
scale_factor_y = 224 / height
|
| 74 |
+
scale_factor_x = 224 / width
|
| 75 |
+
|
| 76 |
+
'''rescale and retrieve landmarks'''
|
| 77 |
+
landmark_arr_xy, landmark_arr_x, landmark_arr_y = \
|
| 78 |
+
self.create_landmarks(landmarks=points_arr,
|
| 79 |
+
scale_factor_x=scale_factor_x,
|
| 80 |
+
scale_factor_y=scale_factor_y)
|
| 81 |
+
|
| 82 |
+
if not(min(landmark_arr_x) < 0.0 or min(landmark_arr_y) < 0.0 or max(landmark_arr_x) > 224 or max(landmark_arr_y) > 224):
|
| 83 |
+
|
| 84 |
+
# self.print_image_arr(fn, resized_img, landmark_arr_x, landmark_arr_y)
|
| 85 |
+
|
| 86 |
+
im = Image.fromarray((resized_img * 255).astype(np.uint8))
|
| 87 |
+
im.save(str(file_name) + '.jpg')
|
| 88 |
+
|
| 89 |
+
pnt_file = open(str(file_name) + ".pts", "w")
|
| 90 |
+
pre_txt = ["version: 1 \n", "n_points: 68 \n", "{ \n"]
|
| 91 |
+
pnt_file.writelines(pre_txt)
|
| 92 |
+
points_txt = ""
|
| 93 |
+
for i in range(0, len(landmark_arr_xy), 2):
|
| 94 |
+
points_txt += str(landmark_arr_xy[i]) + " " + str(landmark_arr_xy[i + 1]) + "\n"
|
| 95 |
+
|
| 96 |
+
pnt_file.writelines(points_txt)
|
| 97 |
+
pnt_file.write("} \n")
|
| 98 |
+
pnt_file.close()
|
| 99 |
+
|
| 100 |
+
return t_label, output_img
|
| 101 |
+
|
| 102 |
+
|
| 103 |
+
|
| 104 |
+
def random_rotate_m(self, _image, _label_img, file_name):
|
| 105 |
+
|
| 106 |
+
rot = random.uniform(-80.9, 80.9)
|
| 107 |
+
|
| 108 |
+
output_img = rotate(_image, rot, resize=True)
|
| 109 |
+
output_img_lbl = rotate(_label_img, rot, resize=True)
|
| 110 |
+
|
| 111 |
+
im = Image.fromarray((output_img * 255).astype(np.uint8))
|
| 112 |
+
im_lbl = Image.fromarray((output_img_lbl * 255).astype(np.uint8))
|
| 113 |
+
|
| 114 |
+
im_m = ImageOps.mirror(im)
|
| 115 |
+
im_lbl_m = ImageOps.mirror(im_lbl)
|
| 116 |
+
|
| 117 |
+
im.save(str(file_name)+'.jpg')
|
| 118 |
+
# im_lbl.save(str(file_name)+'_lbl.jpg')
|
| 119 |
+
|
| 120 |
+
im_m.save(str(file_name) + '_m.jpg')
|
| 121 |
+
# im_lbl_m.save(str(file_name) + '_m_lbl.jpg')
|
| 122 |
+
|
| 123 |
+
im_lbl_ar = np.array(im_lbl)
|
| 124 |
+
im_lbl_m_ar = np.array(im_lbl_m)
|
| 125 |
+
|
| 126 |
+
self.__save_label(im_lbl_ar, file_name, np.array(im))
|
| 127 |
+
self.__save_label(im_lbl_m_ar, file_name+"_m", np.array(im_m))
|
| 128 |
+
|
| 129 |
+
|
| 130 |
+
def __save_label(self, im_lbl_ar, file_name, img_arr):
|
| 131 |
+
|
| 132 |
+
im_lbl_point = []
|
| 133 |
+
for i in range(im_lbl_ar.shape[0]):
|
| 134 |
+
for j in range(im_lbl_ar.shape[1]):
|
| 135 |
+
if im_lbl_ar[i, j] != 0:
|
| 136 |
+
im_lbl_point.append(j)
|
| 137 |
+
im_lbl_point.append(i)
|
| 138 |
+
|
| 139 |
+
pnt_file = open(str(file_name)+".pts", "w")
|
| 140 |
+
|
| 141 |
+
pre_txt = ["version: 1 \n", "n_points: 68 \n", "{ \n"]
|
| 142 |
+
pnt_file.writelines(pre_txt)
|
| 143 |
+
points_txt = ""
|
| 144 |
+
for i in range(0, len(im_lbl_point), 2):
|
| 145 |
+
points_txt += str(im_lbl_point[i]) + " " + str(im_lbl_point[i+1]) + "\n"
|
| 146 |
+
|
| 147 |
+
pnt_file.writelines(points_txt)
|
| 148 |
+
pnt_file.write("} \n")
|
| 149 |
+
pnt_file.close()
|
| 150 |
+
|
| 151 |
+
'''crop data: we add a small margin to the images'''
|
| 152 |
+
xy_points, x_points, y_points = self.create_landmarks(landmarks=im_lbl_point,
|
| 153 |
+
scale_factor_x=1, scale_factor_y=1)
|
| 154 |
+
img_arr, points_arr = self.cropImg(img_arr, x_points, y_points)
|
| 155 |
+
|
| 156 |
+
'''resize image to 224*224'''
|
| 157 |
+
resized_img = resize(img_arr,
|
| 158 |
+
(224, 224, 3),
|
| 159 |
+
anti_aliasing=True)
|
| 160 |
+
dims = img_arr.shape
|
| 161 |
+
height = dims[0]
|
| 162 |
+
width = dims[1]
|
| 163 |
+
scale_factor_y = 224 / height
|
| 164 |
+
scale_factor_x = 224 / width
|
| 165 |
+
|
| 166 |
+
'''rescale and retrieve landmarks'''
|
| 167 |
+
landmark_arr_xy, landmark_arr_x, landmark_arr_y = \
|
| 168 |
+
self.create_landmarks(landmarks=points_arr,
|
| 169 |
+
scale_factor_x=scale_factor_x,
|
| 170 |
+
scale_factor_y=scale_factor_y)
|
| 171 |
+
|
| 172 |
+
im = Image.fromarray((resized_img * 255).astype(np.uint8))
|
| 173 |
+
im.save(str(im_lbl_point[0])+'.jpg')
|
| 174 |
+
# self.print_image_arr(im_lbl_point[0], resized_img, landmark_arr_x, landmark_arr_y)
|
| 175 |
+
|
| 176 |
+
|
| 177 |
+
def augment(self, _image, _label):
|
| 178 |
+
|
| 179 |
+
# face = misc.face(gray=True)
|
| 180 |
+
#
|
| 181 |
+
# rotate_face = ndimage.rotate(_image, 45)
|
| 182 |
+
# self.print_image_arr(_label[0], rotate_face, [],[])
|
| 183 |
+
|
| 184 |
+
# hue_img = tf.image.random_hue(_image, max_delta=0.1) # max_delta must be in the interval [0, 0.5].
|
| 185 |
+
# sat_img = tf.image.random_saturation(hue_img, lower=0.0, upper=3.0)
|
| 186 |
+
#
|
| 187 |
+
# sat_img = K.eval(sat_img)
|
| 188 |
+
#
|
| 189 |
+
_image = self.__noisy(_image)
|
| 190 |
+
|
| 191 |
+
shear = 0
|
| 192 |
+
|
| 193 |
+
# rot = 0.0
|
| 194 |
+
# scale = (1, 1)
|
| 195 |
+
|
| 196 |
+
rot = np.random.uniform(-1 * 0.009, 0.009)
|
| 197 |
+
|
| 198 |
+
scale = (random.uniform(0.9, 1.00), random.uniform(0.9, 1.00))
|
| 199 |
+
|
| 200 |
+
tform = AffineTransform(scale=scale, rotation=rot, shear=shear,
|
| 201 |
+
translation=(0, 0))
|
| 202 |
+
|
| 203 |
+
output_img = warp(_image, tform.inverse, output_shape=(_image.shape[0], _image.shape[1]))
|
| 204 |
+
|
| 205 |
+
sx, sy = scale
|
| 206 |
+
t_matrix = np.array([
|
| 207 |
+
[sx * math.cos(rot), -sy * math.sin(rot + shear), 0],
|
| 208 |
+
[sx * math.sin(rot), sy * math.cos(rot + shear), 0],
|
| 209 |
+
[0, 0, 1]
|
| 210 |
+
])
|
| 211 |
+
landmark_arr_xy, landmark_arr_x, landmark_arr_y = self.create_landmarks(_label, 1, 1)
|
| 212 |
+
label = np.array(landmark_arr_x + landmark_arr_y).reshape([2, 68])
|
| 213 |
+
marging = np.ones([1, 68])
|
| 214 |
+
label = np.concatenate((label, marging), axis=0)
|
| 215 |
+
|
| 216 |
+
label_t = np.dot(t_matrix, label)
|
| 217 |
+
lbl_flat = np.delete(label_t, 2, axis=0).reshape([136])
|
| 218 |
+
|
| 219 |
+
t_label = self.__reorder(lbl_flat)
|
| 220 |
+
return t_label, output_img
|
| 221 |
+
|
| 222 |
+
def __noisy(self, image):
|
| 223 |
+
noise_typ = random.randint(0, 3)
|
| 224 |
+
if noise_typ == 0 :#"gauss":
|
| 225 |
+
row, col, ch = image.shape
|
| 226 |
+
mean = 0
|
| 227 |
+
var = 0.1
|
| 228 |
+
sigma = var ** 0.5
|
| 229 |
+
gauss = np.random.normal(mean, sigma, (row, col, ch))
|
| 230 |
+
gauss = gauss.reshape(row, col, ch)
|
| 231 |
+
noisy = image + gauss
|
| 232 |
+
return noisy
|
| 233 |
+
elif noise_typ == 1 :# "s&p":
|
| 234 |
+
row, col, ch = image.shape
|
| 235 |
+
s_vs_p = 0.5
|
| 236 |
+
amount = 0.004
|
| 237 |
+
out = np.copy(image)
|
| 238 |
+
# Salt mode
|
| 239 |
+
num_salt = np.ceil(amount * image.size * s_vs_p)
|
| 240 |
+
coords = [np.random.randint(0, i - 1, int(num_salt))
|
| 241 |
+
for i in image.shape]
|
| 242 |
+
out[coords] = 1
|
| 243 |
+
|
| 244 |
+
# Pepper mode
|
| 245 |
+
num_pepper = np.ceil(amount * image.size * (1. - s_vs_p))
|
| 246 |
+
coords = [np.random.randint(0, i - 1, int(num_pepper))
|
| 247 |
+
for i in image.shape]
|
| 248 |
+
out[coords] = 0
|
| 249 |
+
return out
|
| 250 |
+
|
| 251 |
+
elif noise_typ == 2: #"speckle":
|
| 252 |
+
row, col, ch = image.shape
|
| 253 |
+
gauss = np.random.randn(row, col, ch)
|
| 254 |
+
gauss = gauss.reshape(row, col, ch)
|
| 255 |
+
noisy = image + image * gauss
|
| 256 |
+
return noisy
|
| 257 |
+
else:
|
| 258 |
+
return image
|
| 259 |
+
|
| 260 |
+
def __reorder(self, input_arr):
|
| 261 |
+
out_arr = []
|
| 262 |
+
for i in range(68):
|
| 263 |
+
out_arr.append(input_arr[i])
|
| 264 |
+
k = 68 + i
|
| 265 |
+
out_arr.append(input_arr[k])
|
| 266 |
+
return np.array(out_arr)
|
| 267 |
+
|
| 268 |
+
def print_image_arr_heat(self, k, image):
|
| 269 |
+
plt.figure()
|
| 270 |
+
plt.imshow(image)
|
| 271 |
+
implot = plt.imshow(image)
|
| 272 |
+
plt.axis('off')
|
| 273 |
+
plt.savefig('heat' + str(k) + '.png', bbox_inches='tight')
|
| 274 |
+
plt.clf()
|
| 275 |
+
|
| 276 |
+
def print_image_arr(self, k, image, landmarks_x, landmarks_y):
|
| 277 |
+
plt.figure()
|
| 278 |
+
plt.imshow(image)
|
| 279 |
+
implot = plt.imshow(image)
|
| 280 |
+
|
| 281 |
+
plt.scatter(x=landmarks_x[:], y=landmarks_y[:], c='black', s=20)
|
| 282 |
+
plt.scatter(x=landmarks_x[:], y=landmarks_y[:], c='white', s=15)
|
| 283 |
+
plt.axis('off')
|
| 284 |
+
plt.savefig('sss' + str(k) + '.png', bbox_inches='tight')
|
| 285 |
+
# plt.show()
|
| 286 |
+
plt.clf()
|
| 287 |
+
|
| 288 |
+
def create_landmarks_from_normalized_original_img(self, img, landmarks, width, height, x_center, y_center, x1, y1, scale_x, scale_y):
|
| 289 |
+
# landmarks_splited = _landmarks.split(';')
|
| 290 |
+
landmark_arr_xy = []
|
| 291 |
+
landmark_arr_x = []
|
| 292 |
+
landmark_arr_y = []
|
| 293 |
+
|
| 294 |
+
for j in range(0, len(landmarks), 2):
|
| 295 |
+
x = ((x_center - float(landmarks[j]) * width)*scale_x) + x1
|
| 296 |
+
y = ((y_center - float(landmarks[j + 1]) * height)*scale_y) + y1
|
| 297 |
+
|
| 298 |
+
landmark_arr_xy.append(x)
|
| 299 |
+
landmark_arr_xy.append(y)
|
| 300 |
+
|
| 301 |
+
landmark_arr_x.append(x)
|
| 302 |
+
landmark_arr_y.append(y)
|
| 303 |
+
|
| 304 |
+
img = cv2.circle(img, (int(x), int(y)), 2, (255, 14, 74), 2)
|
| 305 |
+
img = cv2.circle(img, (int(x), int(y)), 1, (0, 255, 255), 1)
|
| 306 |
+
|
| 307 |
+
return landmark_arr_xy, landmark_arr_x, landmark_arr_y, img
|
| 308 |
+
|
| 309 |
+
|
| 310 |
+
def create_landmarks_from_normalized(self, landmarks, width, height, x_center, y_center):
|
| 311 |
+
|
| 312 |
+
# landmarks_splited = _landmarks.split(';')
|
| 313 |
+
landmark_arr_xy = []
|
| 314 |
+
landmark_arr_x = []
|
| 315 |
+
landmark_arr_y = []
|
| 316 |
+
|
| 317 |
+
for j in range(0, len(landmarks), 2):
|
| 318 |
+
x = x_center - float(landmarks[j]) * width
|
| 319 |
+
y = y_center - float(landmarks[j + 1]) * height
|
| 320 |
+
|
| 321 |
+
landmark_arr_xy.append(x)
|
| 322 |
+
landmark_arr_xy.append(y) # [ x1, y1, x2,y2 ]
|
| 323 |
+
|
| 324 |
+
landmark_arr_x.append(x) # [x1, x2]
|
| 325 |
+
landmark_arr_y.append(y) # [y1, y2]
|
| 326 |
+
|
| 327 |
+
return landmark_arr_xy, landmark_arr_x, landmark_arr_y
|
| 328 |
+
|
| 329 |
+
def create_landmarks(self, landmarks, scale_factor_x=1, scale_factor_y=1):
|
| 330 |
+
# landmarks_splited = _landmarks.split(';')
|
| 331 |
+
landmark_arr_xy = []
|
| 332 |
+
landmark_arr_x = []
|
| 333 |
+
landmark_arr_y = []
|
| 334 |
+
for j in range(0, len(landmarks), 2):
|
| 335 |
+
|
| 336 |
+
x = float(landmarks[j]) * scale_factor_x
|
| 337 |
+
y = float(landmarks[j + 1]) * scale_factor_y
|
| 338 |
+
|
| 339 |
+
landmark_arr_xy.append(x)
|
| 340 |
+
landmark_arr_xy.append(y) # [ x1, y1, x2,y2 ]
|
| 341 |
+
|
| 342 |
+
landmark_arr_x.append(x) # [x1, x2]
|
| 343 |
+
landmark_arr_y.append(y) # [y1, y2]
|
| 344 |
+
|
| 345 |
+
return landmark_arr_xy, landmark_arr_x, landmark_arr_y
|
| 346 |
+
|
| 347 |
+
def create_landmarks_aflw(self, landmarks, scale_factor_x, scale_factor_y):
|
| 348 |
+
# landmarks_splited = _landmarks.split(';')
|
| 349 |
+
landmark_arr_xy = []
|
| 350 |
+
landmark_arr_x = []
|
| 351 |
+
landmark_arr_y = []
|
| 352 |
+
for j in range(0, len(landmarks), 2):
|
| 353 |
+
if landmarks[j][0] == 1:
|
| 354 |
+
x = float(landmarks[j][1]) * scale_factor_x
|
| 355 |
+
y = float(landmarks[j][2]) * scale_factor_y
|
| 356 |
+
|
| 357 |
+
landmark_arr_xy.append(x)
|
| 358 |
+
landmark_arr_xy.append(y) # [ x1, y1, x2,y2 ]
|
| 359 |
+
|
| 360 |
+
landmark_arr_x.append(x) # [x1, x2]
|
| 361 |
+
landmark_arr_y.append(y) # [y1, y2]
|
| 362 |
+
|
| 363 |
+
return landmark_arr_xy, landmark_arr_x, landmark_arr_y
|
| 364 |
+
|
| 365 |
+
def random_augmentation(self, lbl, img):
|
| 366 |
+
# a = random.randint(0, 1)
|
| 367 |
+
# if a == 0:
|
| 368 |
+
# img, lbl = self.__add_margin(img, img.shape[0], lbl)
|
| 369 |
+
|
| 370 |
+
img, lbl = self.__add_margin(img, img.shape[0], lbl)
|
| 371 |
+
|
| 372 |
+
# else:
|
| 373 |
+
# img, lbl = self.__negative_crop(img, lbl)
|
| 374 |
+
|
| 375 |
+
# i = random.randint(0, 2)
|
| 376 |
+
# if i == 0:
|
| 377 |
+
# img, lbl = self.__rotate(img, lbl, 90, img.shape[0], img.shape[1])
|
| 378 |
+
# elif i == 1:
|
| 379 |
+
# img, lbl = self.__rotate(img, lbl, 180, img.shape[0], img.shape[1])
|
| 380 |
+
# else:
|
| 381 |
+
# img, lbl = self.__rotate(img, lbl, 270, img.shape[0], img.shape[1])
|
| 382 |
+
|
| 383 |
+
k = random.randint(0, 3)
|
| 384 |
+
if k > 0:
|
| 385 |
+
img = self.__change_color(img)
|
| 386 |
+
|
| 387 |
+
lbl = np.reshape(lbl, [136])
|
| 388 |
+
return lbl, img
|
| 389 |
+
|
| 390 |
+
|
| 391 |
+
def cropImg_2time(self, img, x_s, y_s):
|
| 392 |
+
min_x = max(0, int(min(x_s) - 150))
|
| 393 |
+
max_x = int(max(x_s) + 150)
|
| 394 |
+
min_y = max(0, int(min(y_s) - 150))
|
| 395 |
+
max_y = int(max(y_s) + 150)
|
| 396 |
+
|
| 397 |
+
crop = img[min_y:max_y, min_x:max_x]
|
| 398 |
+
|
| 399 |
+
new_x_s = []
|
| 400 |
+
new_y_s = []
|
| 401 |
+
new_xy_s = []
|
| 402 |
+
|
| 403 |
+
for i in range(len(x_s)):
|
| 404 |
+
new_x_s.append(x_s[i] - min_x)
|
| 405 |
+
new_y_s.append(y_s[i] - min_y)
|
| 406 |
+
new_xy_s.append(x_s[i] - min_x)
|
| 407 |
+
new_xy_s.append(y_s[i] - min_y)
|
| 408 |
+
return crop, new_xy_s
|
| 409 |
+
|
| 410 |
+
def cropImg(self, img, x_s, y_s, no_padding=False):
|
| 411 |
+
margin1 = random.randint(5, 20)
|
| 412 |
+
margin2 = random.randint(5, 20)
|
| 413 |
+
margin3 = random.randint(5, 20)
|
| 414 |
+
margin4 = random.randint(5, 20)
|
| 415 |
+
|
| 416 |
+
if no_padding:
|
| 417 |
+
min_x = max(0, int(min(x_s)))
|
| 418 |
+
max_x = int(max(x_s))
|
| 419 |
+
min_y = max(0, int(min(y_s)))
|
| 420 |
+
max_y = int(max(y_s))
|
| 421 |
+
else:
|
| 422 |
+
min_x = max(0, int(min(x_s) - margin1))
|
| 423 |
+
max_x = int(max(x_s) + margin2)
|
| 424 |
+
min_y = max(0, int(min(y_s) - margin3))
|
| 425 |
+
max_y = int(max(y_s) + margin4)
|
| 426 |
+
|
| 427 |
+
crop = img[min_y:max_y, min_x:max_x]
|
| 428 |
+
|
| 429 |
+
new_x_s = []
|
| 430 |
+
new_y_s = []
|
| 431 |
+
new_xy_s = []
|
| 432 |
+
|
| 433 |
+
for i in range(len(x_s)):
|
| 434 |
+
new_x_s.append(x_s[i] - min_x)
|
| 435 |
+
new_y_s.append(y_s[i] - min_y)
|
| 436 |
+
new_xy_s.append(x_s[i] - min_x)
|
| 437 |
+
new_xy_s.append(y_s[i] - min_y)
|
| 438 |
+
|
| 439 |
+
# imgpr.print_image_arr(k, crop, new_x_s, new_y_s)
|
| 440 |
+
# imgpr.print_image_arr_2(i, img, x_s, y_s, [min_x, max_x], [min_y, max_y])
|
| 441 |
+
|
| 442 |
+
return crop, new_xy_s
|
| 443 |
+
|
| 444 |
+
def __negative_crop(self, img, landmarks):
|
| 445 |
+
|
| 446 |
+
landmark_arr_xy, x_s, y_s = self.create_landmarks(landmarks, 1, 1)
|
| 447 |
+
min_x = img.shape[0] // random.randint(5, 15)
|
| 448 |
+
max_x = img.shape[0] - (img.shape[0] // random.randint(15, 20))
|
| 449 |
+
min_y = img.shape[0] // random.randint(5, 15)
|
| 450 |
+
max_y = img.shape[0] - (img.shape[0] // random.randint(15, 20))
|
| 451 |
+
|
| 452 |
+
crop = img[min_y:max_y, min_x:max_x]
|
| 453 |
+
|
| 454 |
+
new_x_s = []
|
| 455 |
+
new_y_s = []
|
| 456 |
+
new_xy_s = []
|
| 457 |
+
|
| 458 |
+
for i in range(len(x_s)):
|
| 459 |
+
new_x_s.append(x_s[i] - min_x)
|
| 460 |
+
new_y_s.append(y_s[i] - min_y)
|
| 461 |
+
new_xy_s.append(x_s[i] - min_x)
|
| 462 |
+
new_xy_s.append(y_s[i] - min_y)
|
| 463 |
+
|
| 464 |
+
# imgpr.print_image_arr(crop.shape[0], crop, new_x_s, new_y_s)
|
| 465 |
+
# imgpr.print_image_arr_2(crop.shape[0], crop, x_s, y_s, [min_x, max_x], [min_y, max_y])
|
| 466 |
+
|
| 467 |
+
return crop, new_xy_s
|
| 468 |
+
|
| 469 |
+
def __add_margin(self, img, img_w, lbl):
|
| 470 |
+
marging_width = img_w // random.randint(15, 20)
|
| 471 |
+
direction = random.randint(0, 4)
|
| 472 |
+
|
| 473 |
+
if direction == 1:
|
| 474 |
+
margings = np.random.random([img_w, int(marging_width), 3])
|
| 475 |
+
img = np.concatenate((img, margings), axis=1)
|
| 476 |
+
|
| 477 |
+
if direction == 2:
|
| 478 |
+
margings_1 = np.random.random([img_w, int(marging_width), 3])
|
| 479 |
+
img = np.concatenate((img, margings_1), axis=1)
|
| 480 |
+
|
| 481 |
+
marging_width_1 = img_w // random.randint(15, 20)
|
| 482 |
+
margings_2 = np.random.random([int(marging_width_1), img_w + int(marging_width), 3])
|
| 483 |
+
img = np.concatenate((img, margings_2), axis=0)
|
| 484 |
+
|
| 485 |
+
if direction == 3: # need chane labels
|
| 486 |
+
margings_1 = np.random.random([img_w, int(marging_width), 3])
|
| 487 |
+
img = np.concatenate((margings_1, img), axis=1)
|
| 488 |
+
lbl = self.__transfer_lbl(int(marging_width), lbl, [1, 0])
|
| 489 |
+
|
| 490 |
+
marging_width_1 = img_w // random.randint(15, 20)
|
| 491 |
+
margings_2 = np.random.random([int(marging_width_1), img_w + int(marging_width), 3])
|
| 492 |
+
img = np.concatenate((margings_2, img), axis=0)
|
| 493 |
+
lbl = self.__transfer_lbl(int(marging_width_1), lbl, [0, 1])
|
| 494 |
+
|
| 495 |
+
if direction == 4: # need chane labels
|
| 496 |
+
margings_1 = np.random.random([img_w, int(marging_width), 3])
|
| 497 |
+
img = np.concatenate((margings_1, img), axis=1)
|
| 498 |
+
lbl = self.__transfer_lbl(int(marging_width), lbl, [1, 0])
|
| 499 |
+
img_w1 = img_w + int(marging_width)
|
| 500 |
+
|
| 501 |
+
marging_width_1 = img_w // random.randint(15, 20)
|
| 502 |
+
margings_2 = np.random.random([int(marging_width_1), img_w1, 3])
|
| 503 |
+
img = np.concatenate((margings_2, img), axis=0)
|
| 504 |
+
lbl = self.__transfer_lbl(int(marging_width_1), lbl, [0, 1])
|
| 505 |
+
img_w2 = img_w + int(marging_width_1)
|
| 506 |
+
|
| 507 |
+
marging_width_1 = img_w // random.randint(15, 20)
|
| 508 |
+
margings_1 = np.random.random([img_w2, int(marging_width_1), 3])
|
| 509 |
+
img = np.concatenate((img, margings_1), axis=1)
|
| 510 |
+
|
| 511 |
+
marging_width_1 = img_w // random.randint(15, 20)
|
| 512 |
+
margings_2 = np.random.random([int(marging_width_1), img.shape[1], 3])
|
| 513 |
+
img = np.concatenate((img, margings_2), axis=0)
|
| 514 |
+
|
| 515 |
+
return img, lbl
|
| 516 |
+
|
| 517 |
+
def __void_image(self, img, img_w, ):
|
| 518 |
+
marging_width = int(img_w / random.randint(7, 16))
|
| 519 |
+
direction = random.randint(0, 1)
|
| 520 |
+
direction = 0
|
| 521 |
+
if direction == 0:
|
| 522 |
+
np.delete(img, 100, 1)
|
| 523 |
+
# img[:, 0:marging_width, :] = 0
|
| 524 |
+
elif direction == 1:
|
| 525 |
+
img[img_w - marging_width:img_w, :, :] = 0
|
| 526 |
+
if direction == 2:
|
| 527 |
+
img[:, img_w - marging_width:img_w, :] = 0
|
| 528 |
+
|
| 529 |
+
return img
|
| 530 |
+
|
| 531 |
+
def __change_color(self, img):
|
| 532 |
+
# color_arr = np.random.random([img.shape[0], img.shape[1]])
|
| 533 |
+
color_arr = np.zeros([img.shape[0], img.shape[1]])
|
| 534 |
+
axis = random.randint(0, 4)
|
| 535 |
+
|
| 536 |
+
if axis == 0: # red
|
| 537 |
+
img_mono = img[:, :, 0]
|
| 538 |
+
new_img = np.stack([img_mono, color_arr, color_arr], axis=2)
|
| 539 |
+
elif axis == 1: # green
|
| 540 |
+
img_mono = img[:, :, 1]
|
| 541 |
+
new_img = np.stack([color_arr, img_mono, color_arr], axis=2)
|
| 542 |
+
elif axis == 2: # blue
|
| 543 |
+
img_mono = img[:, :, 1]
|
| 544 |
+
new_img = np.stack([color_arr, img_mono, color_arr], axis=2)
|
| 545 |
+
elif axis == 3: # gray scale
|
| 546 |
+
img_mono = img[:, :, 0]
|
| 547 |
+
new_img = np.stack([img_mono, img_mono, img_mono], axis=2)
|
| 548 |
+
else: # random noise
|
| 549 |
+
color_arr = np.random.random([img.shape[0], img.shape[1]])
|
| 550 |
+
img_mono = img[:, :, 0]
|
| 551 |
+
new_img = np.stack([img_mono, img_mono, color_arr], axis=2)
|
| 552 |
+
|
| 553 |
+
return new_img
|
| 554 |
+
|
| 555 |
+
def __rotate_origin_only(self, xy_arr, radians, xs, ys):
|
| 556 |
+
"""Only rotate a point around the origin (0, 0)."""
|
| 557 |
+
rotated = []
|
| 558 |
+
for xy in xy_arr:
|
| 559 |
+
x, y = xy
|
| 560 |
+
xx = x * math.cos(radians) + y * math.sin(radians)
|
| 561 |
+
yy = -x * math.sin(radians) + y * math.cos(radians)
|
| 562 |
+
rotated.append([xx + xs, yy + ys])
|
| 563 |
+
return np.array(rotated)
|
| 564 |
+
|
| 565 |
+
def __rotate(self, img, landmark_old, degree, img_w, img_h):
|
| 566 |
+
landmark_old = np.reshape(landmark_old, [68, 2])
|
| 567 |
+
|
| 568 |
+
theta = math.radians(degree)
|
| 569 |
+
|
| 570 |
+
if degree == 90:
|
| 571 |
+
landmark = self.__rotate_origin_only(landmark_old, theta, 0, img_h)
|
| 572 |
+
return np.rot90(img, 3, axes=(-2, 0)), landmark
|
| 573 |
+
elif degree == 180:
|
| 574 |
+
landmark = self.__rotate_origin_only(landmark_old, theta, img_h, img_w)
|
| 575 |
+
return np.rot90(img, 2, axes=(-2, 0)), landmark
|
| 576 |
+
elif degree == 270:
|
| 577 |
+
landmark = self.__rotate_origin_only(landmark_old, theta, img_w, 0)
|
| 578 |
+
return np.rot90(img, 1, axes=(-2, 0)), landmark
|
| 579 |
+
|
| 580 |
+
def __transfer_lbl(self, marging_width_1, lbl, axis_arr):
|
| 581 |
+
new_lbl = []
|
| 582 |
+
for i in range(0, len(lbl), 2):
|
| 583 |
+
new_lbl.append(lbl[i] + marging_width_1 * axis_arr[0])
|
| 584 |
+
new_lbl.append(lbl[i + 1] + marging_width_1 * axis_arr[1])
|
| 585 |
+
return np.array(new_lbl)
|
img_printer.py
ADDED
|
@@ -0,0 +1,81 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
|
| 2 |
+
import matplotlib
|
| 3 |
+
matplotlib.use('agg')
|
| 4 |
+
import matplotlib.pyplot as plt
|
| 5 |
+
from pathlib import Path
|
| 6 |
+
|
| 7 |
+
|
| 8 |
+
def print_image(image_name, landmarks_x, landmarks_y):
|
| 9 |
+
|
| 10 |
+
my_file = Path(image_name)
|
| 11 |
+
if my_file.is_file():
|
| 12 |
+
im = plt.imread(image_name)
|
| 13 |
+
implot = plt.imshow(im)
|
| 14 |
+
|
| 15 |
+
plt.scatter(x=landmarks_x[:], y=landmarks_y[:], c='r', s=10)
|
| 16 |
+
plt.show()
|
| 17 |
+
|
| 18 |
+
|
| 19 |
+
def print_image_arr_heat(k, image, print_single=False):
|
| 20 |
+
import numpy as np
|
| 21 |
+
for i in range(image.shape[2]):
|
| 22 |
+
img = np.sum(image, axis=2)
|
| 23 |
+
if print_single:
|
| 24 |
+
plt.figure()
|
| 25 |
+
plt.imshow(image[:, :, i])
|
| 26 |
+
# implot = plt.imshow(image[:, :, i])
|
| 27 |
+
plt.axis('off')
|
| 28 |
+
plt.savefig('single_heat_' + str(i+(k*100)) + '.png', bbox_inches='tight')
|
| 29 |
+
plt.clf()
|
| 30 |
+
|
| 31 |
+
plt.figure()
|
| 32 |
+
plt.imshow(img, vmin=0, vmax=1)
|
| 33 |
+
plt.axis('off')
|
| 34 |
+
plt.savefig('heat_' + str(k) + '.png', bbox_inches='tight')
|
| 35 |
+
plt.clf()
|
| 36 |
+
|
| 37 |
+
|
| 38 |
+
def print_image_arr(k, image, landmarks_x, landmarks_y):
|
| 39 |
+
plt.figure()
|
| 40 |
+
plt.imshow(image)
|
| 41 |
+
implot = plt.imshow(image)
|
| 42 |
+
|
| 43 |
+
# for i in range(len(landmarks_x)):
|
| 44 |
+
# plt.text(landmarks_x[i], landmarks_y[i], str(i), fontsize=12, c='red',
|
| 45 |
+
# horizontalalignment='center', verticalalignment='center',
|
| 46 |
+
# bbox={'facecolor': 'blue', 'alpha': 0.3, 'pad': 0.0})
|
| 47 |
+
|
| 48 |
+
plt.scatter(x=landmarks_x[:], y=landmarks_y[:], c='#000000', s=15)
|
| 49 |
+
plt.scatter(x=landmarks_x[:], y=landmarks_y[:], c='#fddb3a', s=8)
|
| 50 |
+
# plt.axis('off')
|
| 51 |
+
plt.savefig('z_' + str(k) + '.png', bbox_inches='tight')
|
| 52 |
+
# plt.show()
|
| 53 |
+
plt.clf()
|
| 54 |
+
|
| 55 |
+
|
| 56 |
+
def print_image_arr_2(k, image, landmarks_x, landmarks_y, xs, ys):
|
| 57 |
+
plt.figure()
|
| 58 |
+
plt.imshow(image)
|
| 59 |
+
implot = plt.imshow(image)
|
| 60 |
+
|
| 61 |
+
# plt.scatter(x=landmarks_x[:], y=landmarks_y[:], c='b', s=5)
|
| 62 |
+
# for i in range(68):
|
| 63 |
+
# plt.annotate(str(i), (landmarks_x[i], landmarks_y[i]), fontsize=6)
|
| 64 |
+
|
| 65 |
+
plt.scatter(x=landmarks_x[:], y=landmarks_y[:], c='b', s=5)
|
| 66 |
+
plt.scatter(x=xs, y=ys, c='r', s=5)
|
| 67 |
+
plt.savefig('sss'+str(k)+'.png')
|
| 68 |
+
# plt.show()
|
| 69 |
+
plt.clf()
|
| 70 |
+
|
| 71 |
+
|
| 72 |
+
def print_two_landmarks(image, landmarks_1, landmarks_2):
|
| 73 |
+
plt.figure()
|
| 74 |
+
plt.imshow(image)
|
| 75 |
+
implot = plt.imshow(image)
|
| 76 |
+
|
| 77 |
+
plt.scatter(x=landmarks_1[:68], y=landmarks_1[68:], c='b', s=10)
|
| 78 |
+
plt.scatter(x=landmarks_2[:68], y=landmarks_2[68:], c='r', s=10)
|
| 79 |
+
# plt.savefig('a'+str(landmarks_x[0])+'.png')
|
| 80 |
+
plt.show()
|
| 81 |
+
plt.clf()
|
main.py
ADDED
|
@@ -0,0 +1,45 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
|
| 2 |
+
from configuration import DatasetName, D300wConf, InputDataSize, CofwConf, WflwConf
|
| 3 |
+
from cnn_model import CNNModel
|
| 4 |
+
from pca_utility import PCAUtility
|
| 5 |
+
from image_utility import ImageUtility
|
| 6 |
+
from student_train import StudentTrainer
|
| 7 |
+
from test import Test
|
| 8 |
+
from teacher_trainer import TeacherTrainer
|
| 9 |
+
|
| 10 |
+
if __name__ == '__main__':
|
| 11 |
+
|
| 12 |
+
'''test models'''
|
| 13 |
+
|
| 14 |
+
'''train Teacher Networks'''
|
| 15 |
+
trainer = TeacherTrainer(dataset_name=DatasetName.w300)
|
| 16 |
+
trainer.train(arch='efficientNet',weight_path=None)
|
| 17 |
+
|
| 18 |
+
|
| 19 |
+
'''Training Student Network'''
|
| 20 |
+
'''300W'''
|
| 21 |
+
st_trainer = StudentTrainer(dataset_name=DatasetName.w300, use_augmneted=True)
|
| 22 |
+
st_trainer.train(arch_student='mobileNetV2', weight_path_student=None,
|
| 23 |
+
loss_weight_student=2.0,
|
| 24 |
+
arch_tough_teacher='efficientNet', weight_path_tough_teacher='./models/teachers/ds_300w_ef_tou.h5',
|
| 25 |
+
loss_weight_tough_teacher=1,
|
| 26 |
+
arch_tol_teacher='efficientNet', weight_path_tol_teacher='./models/teachers/ds_300w_ef_tol.h5',
|
| 27 |
+
loss_weight_tol_teacher=1)
|
| 28 |
+
|
| 29 |
+
'''COFW'''
|
| 30 |
+
st_trainer = StudentTrainer(dataset_name=DatasetName.cofw, use_augmneted=True)
|
| 31 |
+
st_trainer.train(arch_student='mobileNetV2', weight_path_student=None,
|
| 32 |
+
loss_weight_student=2.0,
|
| 33 |
+
arch_tough_teacher='efficientNet', weight_path_tough_teacher='./models/teachers/ds_cofw_ef_tou.h5',
|
| 34 |
+
loss_weight_tough_teacher=1,
|
| 35 |
+
arch_tol_teacher='efficientNet', weight_path_tol_teacher='./models/teachers/ds_cofw_ef_tol.h5',
|
| 36 |
+
loss_weight_tol_teacher=1)
|
| 37 |
+
|
| 38 |
+
'''WFLW'''
|
| 39 |
+
st_trainer = StudentTrainer(dataset_name=DatasetName.wflw, use_augmneted=True)
|
| 40 |
+
st_trainer.train(arch_student='mobileNetV2', weight_path_student=None,
|
| 41 |
+
loss_weight_student=2.0,
|
| 42 |
+
arch_tough_teacher='efficientNet', weight_path_tough_teacher='./models/teachers/ds_wflw_ef_tou.h5',
|
| 43 |
+
loss_weight_tough_teacher=1,
|
| 44 |
+
arch_tol_teacher='efficientNet', weight_path_tol_teacher='./models/teachers/ds_wflw_ef_tol.h5',
|
| 45 |
+
loss_weight_tol_teacher=1)
|
models/students/student_Net_300w.h5
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:5fc9c077037f4f7cc3df664b95f1f98397a11a2b298e121dc5e79f46837cb240
|
| 3 |
+
size 10197672
|
models/students/student_Net_COFW.h5
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:f3e8cc30f7f6bc3327cf3e59332552b2a321142376b27f14e3b957ffb94fb1da
|
| 3 |
+
size 9799400
|
models/students/student_Net_WFLW.h5
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:61f3ee0c6188856377bd9e38109fdfa0aa97170086cfbdd2bc3c3f355f0f82cd
|
| 3 |
+
size 10504872
|
pca_utility.py
ADDED
|
@@ -0,0 +1,286 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
from configuration import DatasetName, DatasetType,\
|
| 2 |
+
AffectnetConf, D300wConf, W300Conf, InputDataSize, LearningConfig, CofwConf, WflwConf
|
| 3 |
+
from image_utility import ImageUtility
|
| 4 |
+
from sklearn.decomposition import PCA, IncrementalPCA
|
| 5 |
+
from sklearn.decomposition import TruncatedSVD
|
| 6 |
+
import numpy as np
|
| 7 |
+
import pickle
|
| 8 |
+
import os
|
| 9 |
+
from tqdm import tqdm
|
| 10 |
+
from numpy import save, load
|
| 11 |
+
import math
|
| 12 |
+
from PIL import Image
|
| 13 |
+
|
| 14 |
+
|
| 15 |
+
class PCAUtility:
|
| 16 |
+
eigenvalues_prefix = "_eigenvalues_"
|
| 17 |
+
eigenvectors_prefix = "_eigenvectors_"
|
| 18 |
+
meanvector_prefix = "_meanvector_"
|
| 19 |
+
|
| 20 |
+
def create_pca_from_npy(self, dataset_name, pca_postfix):
|
| 21 |
+
lbl_arr = []
|
| 22 |
+
path = None
|
| 23 |
+
if dataset_name == DatasetName.ibug:
|
| 24 |
+
path = D300wConf.normalized_point # normalized
|
| 25 |
+
# elif dataset_name == DatasetName.cofw:
|
| 26 |
+
# path = CofwConf.normalized_points_npy_dir # normalized
|
| 27 |
+
# elif dataset_name == DatasetName.wflw:
|
| 28 |
+
# path = WflwConf.normalized_points_npy_dir # normalized
|
| 29 |
+
|
| 30 |
+
lbl_arr = []
|
| 31 |
+
for file in tqdm(os.listdir(path)):
|
| 32 |
+
if file.endswith(".npy"):
|
| 33 |
+
npy_file = os.path.join(path, file)
|
| 34 |
+
lbl_arr.append(load(npy_file))
|
| 35 |
+
|
| 36 |
+
lbl_arr = np.array(lbl_arr)
|
| 37 |
+
|
| 38 |
+
print('PCA calculation started')
|
| 39 |
+
|
| 40 |
+
''' no normalization is needed, since we want to generate hm'''
|
| 41 |
+
reduced_lbl_arr, eigenvalues, eigenvectors = self._func_PCA(lbl_arr, pca_postfix)
|
| 42 |
+
mean_lbl_arr = np.mean(lbl_arr, axis=0)
|
| 43 |
+
eigenvectors = eigenvectors.T
|
| 44 |
+
#
|
| 45 |
+
# self.__save_obj(eigenvalues, dataset_name + self.__eigenvalues_prefix + str(pca_postfix))
|
| 46 |
+
# self.__save_obj(eigenvectors, dataset_name + self.__eigenvectors_prefix + str(pca_postfix))
|
| 47 |
+
# self.__save_obj(mean_lbl_arr, dataset_name + self.__meanvector_prefix + str(pca_postfix))
|
| 48 |
+
#
|
| 49 |
+
save('pca_obj/' + dataset_name + self.eigenvalues_prefix + str(pca_postfix), eigenvalues)
|
| 50 |
+
save('pca_obj/' + dataset_name + self.eigenvectors_prefix + str(pca_postfix), eigenvectors)
|
| 51 |
+
save('pca_obj/' + dataset_name + self.meanvector_prefix + str(pca_postfix), mean_lbl_arr)
|
| 52 |
+
|
| 53 |
+
def create_pca_from_points(self, dataset_name, pca_postfix):
|
| 54 |
+
lbl_arr = []
|
| 55 |
+
path = None
|
| 56 |
+
if dataset_name == DatasetName.ibug:
|
| 57 |
+
path = D300wConf.rotated_img_path_prefix # rotated is ok, since advs_aug is the same as rotated
|
| 58 |
+
num_of_landmarks = D300wConf.num_of_landmarks
|
| 59 |
+
elif dataset_name == DatasetName.cofw:
|
| 60 |
+
path = CofwConf.rotated_img_path_prefix
|
| 61 |
+
num_of_landmarks = CofwConf.num_of_landmarks
|
| 62 |
+
elif dataset_name == DatasetName.wflw:
|
| 63 |
+
path = WflwConf.rotated_img_path_prefix
|
| 64 |
+
num_of_landmarks = WflwConf.num_of_landmarks
|
| 65 |
+
|
| 66 |
+
for file in tqdm(os.listdir(path)):
|
| 67 |
+
if file.endswith(".pts"):
|
| 68 |
+
pts_file = os.path.join(path, file)
|
| 69 |
+
|
| 70 |
+
points_arr = []
|
| 71 |
+
with open(pts_file) as fp:
|
| 72 |
+
line = fp.readline()
|
| 73 |
+
cnt = 1
|
| 74 |
+
while line:
|
| 75 |
+
if 3 < cnt <= num_of_landmarks+3:
|
| 76 |
+
x_y_pnt = line.strip()
|
| 77 |
+
x = float(x_y_pnt.split(" ")[0])
|
| 78 |
+
y = float(x_y_pnt.split(" ")[1])
|
| 79 |
+
points_arr.append(x)
|
| 80 |
+
points_arr.append(y)
|
| 81 |
+
line = fp.readline()
|
| 82 |
+
cnt += 1
|
| 83 |
+
lbl_arr.append(points_arr)
|
| 84 |
+
|
| 85 |
+
lbl_arr = np.array(lbl_arr)
|
| 86 |
+
|
| 87 |
+
print('PCA calculation started')
|
| 88 |
+
|
| 89 |
+
''' no normalization is needed, since we want to generate hm'''
|
| 90 |
+
reduced_lbl_arr, eigenvalues, eigenvectors = self._func_PCA(lbl_arr, pca_postfix)
|
| 91 |
+
mean_lbl_arr = np.mean(lbl_arr, axis=0)
|
| 92 |
+
eigenvectors = eigenvectors.T
|
| 93 |
+
#
|
| 94 |
+
# self.__save_obj(eigenvalues, dataset_name + self.__eigenvalues_prefix + str(pca_postfix))
|
| 95 |
+
# self.__save_obj(eigenvectors, dataset_name + self.__eigenvectors_prefix + str(pca_postfix))
|
| 96 |
+
# self.__save_obj(mean_lbl_arr, dataset_name + self.__meanvector_prefix + str(pca_postfix))
|
| 97 |
+
#
|
| 98 |
+
save('pca_obj/' + dataset_name + self.eigenvalues_prefix + str(pca_postfix), eigenvalues)
|
| 99 |
+
save('pca_obj/' + dataset_name + self.eigenvectors_prefix + str(pca_postfix), eigenvectors)
|
| 100 |
+
save('pca_obj/' + dataset_name + self.meanvector_prefix + str(pca_postfix), mean_lbl_arr)
|
| 101 |
+
|
| 102 |
+
def test_pca_validity(self, dataset_name, pca_postfix):
|
| 103 |
+
image_utility = ImageUtility()
|
| 104 |
+
|
| 105 |
+
eigenvalues = load('pca_obj/' + dataset_name + self.eigenvalues_prefix + str(pca_postfix)+".npy")
|
| 106 |
+
eigenvectors = load('pca_obj/' + dataset_name + self.eigenvectors_prefix + str(pca_postfix)+".npy")
|
| 107 |
+
meanvector = load('pca_obj/' + dataset_name + self.meanvector_prefix + str(pca_postfix)+".npy")
|
| 108 |
+
|
| 109 |
+
'''load data: '''
|
| 110 |
+
lbl_arr = []
|
| 111 |
+
img_arr = []
|
| 112 |
+
path = None
|
| 113 |
+
|
| 114 |
+
if dataset_name == DatasetName.ibug:
|
| 115 |
+
path = D300wConf.rotated_img_path_prefix # rotated is ok, since advs_aug is the same as rotated
|
| 116 |
+
num_of_landmarks = D300wConf.num_of_landmarks
|
| 117 |
+
elif dataset_name == DatasetName.cofw:
|
| 118 |
+
path = CofwConf.rotated_img_path_prefix
|
| 119 |
+
num_of_landmarks = CofwConf.num_of_landmarks
|
| 120 |
+
elif dataset_name == DatasetName.wflw:
|
| 121 |
+
path = WflwConf.rotated_img_path_prefix
|
| 122 |
+
num_of_landmarks = WflwConf.num_of_landmarks
|
| 123 |
+
|
| 124 |
+
for file in tqdm(os.listdir(path)):
|
| 125 |
+
if file.endswith(".pts"):
|
| 126 |
+
pts_file = os.path.join(path, file)
|
| 127 |
+
img_file = pts_file[:-3] + "jpg"
|
| 128 |
+
if not os.path.exists(img_file):
|
| 129 |
+
continue
|
| 130 |
+
|
| 131 |
+
points_arr = []
|
| 132 |
+
with open(pts_file) as fp:
|
| 133 |
+
line = fp.readline()
|
| 134 |
+
cnt = 1
|
| 135 |
+
while line:
|
| 136 |
+
if 3 < cnt <= num_of_landmarks+3:
|
| 137 |
+
x_y_pnt = line.strip()
|
| 138 |
+
x = float(x_y_pnt.split(" ")[0])
|
| 139 |
+
y = float(x_y_pnt.split(" ")[1])
|
| 140 |
+
points_arr.append(x)
|
| 141 |
+
points_arr.append(y)
|
| 142 |
+
line = fp.readline()
|
| 143 |
+
cnt += 1
|
| 144 |
+
lbl_arr.append(points_arr)
|
| 145 |
+
img_arr.append(Image.open(img_file))
|
| 146 |
+
|
| 147 |
+
for i in range(30):
|
| 148 |
+
b_vector_p = self.calculate_b_vector(lbl_arr[i], True, eigenvalues, eigenvectors, meanvector)
|
| 149 |
+
lbl_new = meanvector + np.dot(eigenvectors, b_vector_p)
|
| 150 |
+
lbl_new = lbl_new.tolist()
|
| 151 |
+
|
| 152 |
+
labels_true_transformed, landmark_arr_x_t, landmark_arr_y_t = image_utility. \
|
| 153 |
+
create_landmarks(lbl_arr[i], 1, 1)
|
| 154 |
+
|
| 155 |
+
labels_true_transformed_pca, landmark_arr_x_pca, landmark_arr_y_pca = image_utility. \
|
| 156 |
+
create_landmarks(lbl_new, 1, 1)
|
| 157 |
+
|
| 158 |
+
image_utility.print_image_arr(i, img_arr[i], landmark_arr_x_t, landmark_arr_y_t)
|
| 159 |
+
image_utility.print_image_arr(i * 1000, img_arr[i], landmark_arr_x_pca, landmark_arr_y_pca)
|
| 160 |
+
|
| 161 |
+
def calculate_b_vector(self, predicted_vector, correction, eigenvalues, eigenvectors, meanvector):
|
| 162 |
+
tmp1 = predicted_vector - meanvector
|
| 163 |
+
b_vector = np.dot(eigenvectors.T, tmp1)
|
| 164 |
+
|
| 165 |
+
# put b in -3lambda =>
|
| 166 |
+
if correction:
|
| 167 |
+
i = 0
|
| 168 |
+
for b_item in b_vector:
|
| 169 |
+
lambda_i_sqr = 3 * math.sqrt(eigenvalues[i])
|
| 170 |
+
|
| 171 |
+
if b_item > 0:
|
| 172 |
+
b_item = min(b_item, lambda_i_sqr)
|
| 173 |
+
else:
|
| 174 |
+
b_item = max(b_item, -1 * lambda_i_sqr)
|
| 175 |
+
b_vector[i] = b_item
|
| 176 |
+
i += 1
|
| 177 |
+
|
| 178 |
+
return b_vector
|
| 179 |
+
|
| 180 |
+
def create_pca(self, dataset_name, pca_postfix):
|
| 181 |
+
tf_record_util = TFRecordUtility()
|
| 182 |
+
|
| 183 |
+
lbl_arr = []
|
| 184 |
+
pose_arr = []
|
| 185 |
+
if dataset_name == DatasetName.ibug:
|
| 186 |
+
lbl_arr, img_arr, pose_arr = tf_record_util.retrieve_tf_record(D300wConf.tf_train_path,
|
| 187 |
+
D300wConf.sum_of_train_samples,
|
| 188 |
+
only_label=True, only_pose=True)
|
| 189 |
+
lbl_arr = np.array(lbl_arr)
|
| 190 |
+
|
| 191 |
+
print('PCA-retrieved')
|
| 192 |
+
|
| 193 |
+
'''need to be normalized based on the hyper face paper?'''
|
| 194 |
+
|
| 195 |
+
# reduced_lbl_arr, eigenvalues, eigenvectors = self.__svd_func(lbl_arr, pca_postfix)
|
| 196 |
+
reduced_lbl_arr, eigenvalues, eigenvectors = self.__func_PCA(lbl_arr, pca_postfix)
|
| 197 |
+
mean_lbl_arr = np.mean(lbl_arr, axis=0)
|
| 198 |
+
eigenvectors = eigenvectors.T
|
| 199 |
+
|
| 200 |
+
self.__save_obj(eigenvalues, dataset_name + self.eigenvalues_prefix + str(pca_postfix))
|
| 201 |
+
self.__save_obj(eigenvectors, dataset_name + self.eigenvectors_prefix + str(pca_postfix))
|
| 202 |
+
self.__save_obj(mean_lbl_arr, dataset_name + self.meanvector_prefix + str(pca_postfix))
|
| 203 |
+
|
| 204 |
+
'''calculate pose min max'''
|
| 205 |
+
p_1_arr = []
|
| 206 |
+
p_2_arr = []
|
| 207 |
+
p_3_arr = []
|
| 208 |
+
|
| 209 |
+
for p_item in pose_arr:
|
| 210 |
+
p_1_arr.append(p_item[0])
|
| 211 |
+
p_2_arr.append(p_item[1])
|
| 212 |
+
p_3_arr.append(p_item[2])
|
| 213 |
+
|
| 214 |
+
p_1_min = min(p_1_arr)
|
| 215 |
+
p_1_max = max(p_1_arr)
|
| 216 |
+
|
| 217 |
+
p_2_min = min(p_2_arr)
|
| 218 |
+
p_2_max = max(p_2_arr)
|
| 219 |
+
|
| 220 |
+
p_3_min = min(p_3_arr)
|
| 221 |
+
p_3_max = max(p_3_arr)
|
| 222 |
+
|
| 223 |
+
self.__save_obj(p_1_min, 'p_1_min')
|
| 224 |
+
self.__save_obj(p_1_max, 'p_1_max')
|
| 225 |
+
|
| 226 |
+
self.__save_obj(p_2_min, 'p_2_min')
|
| 227 |
+
self.__save_obj(p_2_max, 'p_2_max')
|
| 228 |
+
|
| 229 |
+
self.__save_obj(p_3_min, 'p_3_min')
|
| 230 |
+
self.__save_obj(p_3_max, 'p_3_max')
|
| 231 |
+
|
| 232 |
+
print('PCA-->done')
|
| 233 |
+
|
| 234 |
+
def __save_obj(self, obj, name):
|
| 235 |
+
with open('obj/' + name + '.pkl', 'wb') as f:
|
| 236 |
+
pickle.dump(obj, f, pickle.HIGHEST_PROTOCOL)
|
| 237 |
+
|
| 238 |
+
def load_pose_obj(self):
|
| 239 |
+
with open('obj/p_1_min.pkl', 'rb') as f:
|
| 240 |
+
p_1_min = pickle.load(f)
|
| 241 |
+
with open('obj/p_1_max.pkl', 'rb') as f:
|
| 242 |
+
p_1_max = pickle.load(f)
|
| 243 |
+
|
| 244 |
+
with open('obj/p_2_min.pkl', 'rb') as f:
|
| 245 |
+
p_2_min = pickle.load(f)
|
| 246 |
+
with open('obj/p_2_max.pkl', 'rb') as f:
|
| 247 |
+
p_2_max = pickle.load(f)
|
| 248 |
+
|
| 249 |
+
with open('obj/p_3_min.pkl', 'rb') as f:
|
| 250 |
+
p_3_min = pickle.load(f)
|
| 251 |
+
with open('obj/p_3_max.pkl', 'rb') as f:
|
| 252 |
+
p_3_max = pickle.load(f)
|
| 253 |
+
|
| 254 |
+
return p_1_min, p_1_max, p_2_min, p_2_max, p_3_min, p_3_max
|
| 255 |
+
|
| 256 |
+
|
| 257 |
+
def load_pca_obj(self, dataset_name, pca_postfix=97):
|
| 258 |
+
with open('obj/' + dataset_name + self.eigenvalues_prefix + str(pca_postfix) + '.pkl', 'rb') as f:
|
| 259 |
+
eigenvalues = pickle.load(f)
|
| 260 |
+
with open('obj/' + dataset_name + self.eigenvectors_prefix + str(pca_postfix) + '.pkl', 'rb') as f:
|
| 261 |
+
eigenvectors = pickle.load(f)
|
| 262 |
+
with open('obj/' + dataset_name + self.meanvector_prefix + str(pca_postfix) + '.pkl', 'rb') as f:
|
| 263 |
+
meanvector = pickle.load(f)
|
| 264 |
+
return eigenvalues, eigenvectors, meanvector
|
| 265 |
+
|
| 266 |
+
def _func_PCA(self, input_data, pca_postfix):
|
| 267 |
+
input_data = np.array(input_data)
|
| 268 |
+
pca = PCA(n_components=pca_postfix/100)
|
| 269 |
+
# pca = PCA(n_components=0.98)
|
| 270 |
+
# pca = IncrementalPCA(n_components=50, batch_size=50)
|
| 271 |
+
pca.fit(input_data)
|
| 272 |
+
pca_input_data = pca.transform(input_data)
|
| 273 |
+
eigenvalues = pca.explained_variance_
|
| 274 |
+
eigenvectors = pca.components_
|
| 275 |
+
return pca_input_data, eigenvalues, eigenvectors
|
| 276 |
+
|
| 277 |
+
def __svd_func(self, input_data, pca_postfix):
|
| 278 |
+
svd = TruncatedSVD(n_components=50)
|
| 279 |
+
svd.fit(input_data)
|
| 280 |
+
pca_input_data = svd.transform(input_data)
|
| 281 |
+
eigenvalues = svd.explained_variance_
|
| 282 |
+
eigenvectors = svd.components_
|
| 283 |
+
return pca_input_data, eigenvalues, eigenvectors
|
| 284 |
+
# U, S, VT = svd(input_data)
|
| 285 |
+
|
| 286 |
+
|
requirements.txt
ADDED
|
@@ -0,0 +1,23 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# for cuda 9
|
| 2 |
+
# pip install torch==1.2.0+cu92 torchvision==0.4.0+cu92 -f https://download.pytorch.org/whl/torch_stable.html
|
| 3 |
+
#
|
| 4 |
+
#torch
|
| 5 |
+
#torchvision
|
| 6 |
+
bottleneck
|
| 7 |
+
numpy==1.19.2
|
| 8 |
+
#tensorflow==1.14.0
|
| 9 |
+
tensorflow==2.3.1
|
| 10 |
+
#tensorflow-gpu==1.14
|
| 11 |
+
# keras==2.2.4
|
| 12 |
+
keras==2.4.3
|
| 13 |
+
matplotlib
|
| 14 |
+
opencv-python
|
| 15 |
+
opencv-contrib-python
|
| 16 |
+
scipy
|
| 17 |
+
scikit-learn
|
| 18 |
+
scikit-image
|
| 19 |
+
Pillow
|
| 20 |
+
tqdm
|
| 21 |
+
efficientnet
|
| 22 |
+
# tfkerassurgeon
|
| 23 |
+
tensorboard
|
samples/KDLoss-1.jpg
ADDED

|
Git LFS Details
|
samples/KD_300W_samples-1.jpg
ADDED

|
Git LFS Details
|
samples/KD_L2_L1-1.jpg
ADDED

|
Git LFS Details
|
samples/KD_WFLW_samples-1.jpg
ADDED

|
Git LFS Details
|
samples/KD_cofw_samples-1.jpg
ADDED

|
Git LFS Details
|
samples/general_framework-1.jpg
ADDED

|
Git LFS Details
|
samples/loss_weight-1.jpg
ADDED

|
Git LFS Details
|
samples/main_loss-1.jpg
ADDED

|
Git LFS Details
|
samples/teacher_arch-1.jpg
ADDED

|
Git LFS Details
|
student_train.py
ADDED
|
@@ -0,0 +1,292 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
from configuration import DatasetName, DatasetType, \
|
| 2 |
+
AffectnetConf, D300wConf, W300Conf, InputDataSize, LearningConfig, CofwConf, WflwConf
|
| 3 |
+
from tf_record_utility import TFRecordUtility
|
| 4 |
+
from clr_callback import CyclicLR
|
| 5 |
+
from cnn_model import CNNModel
|
| 6 |
+
from custom_Losses import Custom_losses
|
| 7 |
+
from Data_custom_generator import CustomHeatmapGenerator
|
| 8 |
+
from PW_Data_custom_generator import PWCustomHeatmapGenerator
|
| 9 |
+
from image_utility import ImageUtility
|
| 10 |
+
import img_printer as imgpr
|
| 11 |
+
|
| 12 |
+
import tensorflow as tf
|
| 13 |
+
import numpy as np
|
| 14 |
+
import matplotlib.pyplot as plt
|
| 15 |
+
import math
|
| 16 |
+
from datetime import datetime
|
| 17 |
+
from sklearn.utils import shuffle
|
| 18 |
+
from sklearn.model_selection import train_test_split
|
| 19 |
+
from numpy import save, load, asarray
|
| 20 |
+
import csv
|
| 21 |
+
from skimage.io import imread
|
| 22 |
+
import pickle
|
| 23 |
+
|
| 24 |
+
|
| 25 |
+
# tf.compat.v1.enable_eager_execution()
|
| 26 |
+
|
| 27 |
+
|
| 28 |
+
class StudentTrainer:
|
| 29 |
+
|
| 30 |
+
def __init__(self, dataset_name, use_augmneted):
|
| 31 |
+
self.dataset_name = dataset_name
|
| 32 |
+
|
| 33 |
+
if dataset_name == DatasetName.w300:
|
| 34 |
+
self.num_landmark = D300wConf.num_of_landmarks * 2
|
| 35 |
+
if use_augmneted:
|
| 36 |
+
self.img_path = D300wConf.augmented_train_image
|
| 37 |
+
self.annotation_path = D300wConf.augmented_train_annotation
|
| 38 |
+
else:
|
| 39 |
+
self.img_path = D300wConf.no_aug_train_image
|
| 40 |
+
self.annotation_path = D300wConf.no_aug_train_annotation
|
| 41 |
+
|
| 42 |
+
if dataset_name == DatasetName.cofw:
|
| 43 |
+
self.num_landmark = CofwConf.num_of_landmarks * 2
|
| 44 |
+
self.img_path = CofwConf.augmented_train_image
|
| 45 |
+
self.annotation_path = CofwConf.augmented_train_annotation
|
| 46 |
+
|
| 47 |
+
if dataset_name == DatasetName.wflw:
|
| 48 |
+
self.num_landmark = WflwConf.num_of_landmarks * 2
|
| 49 |
+
if use_augmneted:
|
| 50 |
+
self.img_path = WflwConf.augmented_train_image
|
| 51 |
+
self.annotation_path = WflwConf.augmented_train_annotation
|
| 52 |
+
else:
|
| 53 |
+
self.img_path = WflwConf.no_aug_train_image
|
| 54 |
+
self.annotation_path = WflwConf.no_aug_train_annotation
|
| 55 |
+
|
| 56 |
+
def train(self, arch_student, weight_path_student, loss_weight_student,
|
| 57 |
+
arch_tough_teacher, weight_path_tough_teacher, loss_weight_tough_teacher,
|
| 58 |
+
arch_tol_teacher, weight_path_tol_teacher, loss_weight_tol_teacher):
|
| 59 |
+
""""""
|
| 60 |
+
'''create loss'''
|
| 61 |
+
c_loss = Custom_losses()
|
| 62 |
+
|
| 63 |
+
'''create summary writer'''
|
| 64 |
+
summary_writer = tf.summary.create_file_writer(
|
| 65 |
+
"./train_logs/fit/" + datetime.now().strftime("%Y%m%d-%H%M%S"))
|
| 66 |
+
|
| 67 |
+
'''making models'''
|
| 68 |
+
model_student = self.make_model(arch=arch_student, w_path=weight_path_student, is_old=False)
|
| 69 |
+
model_tough_teacher = self.make_model(arch=arch_tough_teacher, w_path=weight_path_tough_teacher)
|
| 70 |
+
model_tol_teacher = self.make_model(arch=arch_tol_teacher, w_path=weight_path_tol_teacher)
|
| 71 |
+
|
| 72 |
+
'''create optimizer'''
|
| 73 |
+
_lr = 1e-3
|
| 74 |
+
optimizer_student = self._get_optimizer(lr=_lr)
|
| 75 |
+
|
| 76 |
+
'''create sample generator'''
|
| 77 |
+
x_train_filenames, x_val_filenames, y_train_filenames, y_val_filenames = self._create_generators()
|
| 78 |
+
# x_train_filenames, y_train_filenames = self._create_generators()
|
| 79 |
+
|
| 80 |
+
'''create train configuration'''
|
| 81 |
+
step_per_epoch = len(x_train_filenames) // LearningConfig.batch_size
|
| 82 |
+
|
| 83 |
+
'''start train:'''
|
| 84 |
+
for epoch in range(LearningConfig.epochs):
|
| 85 |
+
x_train_filenames, y_train_filenames = self._shuffle_data(x_train_filenames, y_train_filenames)
|
| 86 |
+
for batch_index in range(step_per_epoch):
|
| 87 |
+
'''load annotation and images'''
|
| 88 |
+
images, annotation_gr, annotation_tough_teacher, annotation_tol_teacher, annotation_student = self._get_batch_sample(
|
| 89 |
+
batch_index=batch_index, x_train_filenames=x_train_filenames,
|
| 90 |
+
y_train_filenames=y_train_filenames, model_tough_t=model_tough_teacher,
|
| 91 |
+
model_tol_t=model_tol_teacher, model_student=model_student)
|
| 92 |
+
'''convert to tensor'''
|
| 93 |
+
images = tf.cast(images, tf.float32)
|
| 94 |
+
annotation_gr = tf.cast(annotation_gr, tf.float32)
|
| 95 |
+
annotation_tough_teacher = tf.cast(annotation_tough_teacher, tf.float32)
|
| 96 |
+
annotation_tol_teacher = tf.cast(annotation_tol_teacher, tf.float32)
|
| 97 |
+
|
| 98 |
+
'''train step'''
|
| 99 |
+
self.train_step(epoch=epoch, step=batch_index, total_steps=step_per_epoch, images=images,
|
| 100 |
+
model_student=model_student,
|
| 101 |
+
annotation_gr=annotation_gr, annotation_tough_teacher=annotation_tough_teacher,
|
| 102 |
+
annotation_tol_teacher=annotation_tol_teacher,
|
| 103 |
+
l_w_stu=loss_weight_student, l_w_togh_t=loss_weight_tough_teacher,
|
| 104 |
+
loss_w_tol_t=loss_weight_tol_teacher,
|
| 105 |
+
optimizer=optimizer_student, summary_writer=summary_writer, c_loss=c_loss)
|
| 106 |
+
'''evaluating part'''
|
| 107 |
+
img_batch_eval, pn_batch_eval = self._create_evaluation_batch(x_val_filenames, y_val_filenames)
|
| 108 |
+
# loss_eval, loss_eval_tol_dif_stu, loss_eval_tol_dif_gt, loss_eval_tou_dif_stu, loss_eval_tou_dif_gt = \
|
| 109 |
+
loss_eval = self._eval_model(img_batch_eval, pn_batch_eval, model_student)
|
| 110 |
+
with summary_writer.as_default():
|
| 111 |
+
tf.summary.scalar('Eval-LOSS', loss_eval, step=epoch)
|
| 112 |
+
# tf.summary.scalar('Eval-loss_eval_tol_dif_stu', loss_eval_tol_dif_stu, step=epoch)
|
| 113 |
+
# tf.summary.scalar('Eval-loss_eval_tol_dif_gt', loss_eval_tol_dif_gt, step=epoch)
|
| 114 |
+
# tf.summary.scalar('Eval-loss_eval_tou_dif_stu', loss_eval_tou_dif_stu, step=epoch)
|
| 115 |
+
# tf.summary.scalar('Eval-loss_eval_tou_dif_gt', loss_eval_tou_dif_gt, step=epoch)
|
| 116 |
+
# model_student.save_weights('./models/stu_weight_' + '_' + str(epoch) + self.dataset_name + '_' + str(loss_eval) + '.h5')
|
| 117 |
+
'''save weights'''
|
| 118 |
+
model_student.save(
|
| 119 |
+
'./models/stu_model_' + str(epoch) + '_' + self.dataset_name + '_' + str(loss_eval) + '.h5')
|
| 120 |
+
'''calculate Learning rate'''
|
| 121 |
+
# _lr = self._calc_learning_rate(iterations=epoch, step_size=20, base_lr=1e-5, max_lr=1e-1)
|
| 122 |
+
# optimizer_student = self._get_optimizer(lr=_lr)
|
| 123 |
+
|
| 124 |
+
def _calc_learning_rate(self, iterations, step_size, base_lr, max_lr):
|
| 125 |
+
cycle = np.floor(1 + iterations / (2 * step_size))
|
| 126 |
+
x = np.abs(iterations / step_size - 2 * cycle + 1)
|
| 127 |
+
lr = base_lr + (max_lr - base_lr) * np.maximum(0, (1 - x)) / float(2 ** (cycle - 1))
|
| 128 |
+
print('LR is: ' + str(lr))
|
| 129 |
+
return lr
|
| 130 |
+
|
| 131 |
+
# @tf.function
|
| 132 |
+
def train_step(self, epoch, step, total_steps, images, model_student, annotation_gr,
|
| 133 |
+
annotation_tough_teacher, annotation_tol_teacher, annotation_student,
|
| 134 |
+
l_w_stu, l_w_togh_t, loss_w_tol_t,
|
| 135 |
+
optimizer, summary_writer, c_loss, train_dif):
|
| 136 |
+
with tf.GradientTape() as tape_student:
|
| 137 |
+
'''create annotation_predicted'''
|
| 138 |
+
# annotation_predicted, pr_tol, pr_tol_dif_gt, pr_tou, pr_tou_dif_gt = model_student(
|
| 139 |
+
annotation_predicted = model_student(
|
| 140 |
+
images, training=True)
|
| 141 |
+
'''calculate loss'''
|
| 142 |
+
loss_total, loss_main, loss_tough_assist, loss_tol_assist = c_loss.kd_loss(x_pr=annotation_predicted,
|
| 143 |
+
x_gt=annotation_gr,
|
| 144 |
+
x_tough=annotation_tough_teacher,
|
| 145 |
+
x_tol=annotation_tol_teacher,
|
| 146 |
+
alpha_tough=1.9,
|
| 147 |
+
alpha_mi_tough=-0.45,
|
| 148 |
+
alpha_tol=1.8,
|
| 149 |
+
alpha_mi_tol=-0.4,
|
| 150 |
+
main_loss_weight=l_w_stu,
|
| 151 |
+
tough_loss_weight=l_w_togh_t,
|
| 152 |
+
tol_loss_weight=loss_w_tol_t)
|
| 153 |
+
'''calculate gradient'''
|
| 154 |
+
gradients_of_student = tape_student.gradient(loss_total, model_student.trainable_variables)
|
| 155 |
+
'''apply Gradients:'''
|
| 156 |
+
optimizer.apply_gradients(zip(gradients_of_student, model_student.trainable_variables))
|
| 157 |
+
'''printing loss Values: '''
|
| 158 |
+
tf.print("->EPOCH: ", str(epoch), "->STEP: ", str(step) + '/' + str(total_steps),
|
| 159 |
+
' -> : LOSS: ', loss_total,
|
| 160 |
+
' -> : loss_main: ', loss_main,
|
| 161 |
+
' -> : loss_tough_assist: ', loss_tough_assist,
|
| 162 |
+
' -> : loss_tol_assist: ', loss_tol_assist)
|
| 163 |
+
with summary_writer.as_default():
|
| 164 |
+
tf.summary.scalar('LOSS', loss_total, step=epoch)
|
| 165 |
+
tf.summary.scalar('loss_main', loss_main, step=epoch)
|
| 166 |
+
tf.summary.scalar('loss_tough_assist', loss_tough_assist, step=epoch)
|
| 167 |
+
tf.summary.scalar('loss_tol_assist', loss_tol_assist, step=epoch)
|
| 168 |
+
|
| 169 |
+
def make_model(self, arch, w_path, is_old=False):
|
| 170 |
+
cnn = CNNModel()
|
| 171 |
+
model = cnn.get_model(arch=arch, output_len=self.num_landmark, input_tensor=None, weight_path=w_path,
|
| 172 |
+
is_old=is_old)
|
| 173 |
+
if w_path is not None and arch != 'mobileNetV2_d' and not is_old:
|
| 174 |
+
model.load_weights(w_path)
|
| 175 |
+
# model.save('test_model'+arch+'.h5')
|
| 176 |
+
return model
|
| 177 |
+
|
| 178 |
+
def _eval_model(self, img_batch_eval, pn_batch_eval, model):
|
| 179 |
+
# annotation_predicted, pr_tol_dif_stu, pr_tol_dif_gt, pr_tou_dif_stu, pr_tou_dif_gt = model(img_batch_eval)
|
| 180 |
+
annotation_predicted = model(img_batch_eval)
|
| 181 |
+
loss_eval = np.array(tf.reduce_mean(tf.abs(pn_batch_eval - annotation_predicted)))
|
| 182 |
+
# loss_eval_tol_dif_stu = np.array(tf.reduce_mean(tf.abs(pn_batch_eval - annotation_predicted)))
|
| 183 |
+
# loss_eval_tol_dif_gt = np.array(tf.reduce_mean(tf.abs(pn_batch_eval - annotation_predicted)))
|
| 184 |
+
# loss_eval_tou_dif_stu = np.array(tf.reduce_mean(tf.abs(pn_batch_eval - annotation_predicted)))
|
| 185 |
+
# loss_eval_tou_dif_gt = np.array(tf.reduce_mean(tf.abs(pn_batch_eval - annotation_predicted)))
|
| 186 |
+
# return loss_eval, loss_eval_tol_dif_stu, loss_eval_tol_dif_gt, loss_eval_tou_dif_stu, loss_eval_tou_dif_gt
|
| 187 |
+
return loss_eval
|
| 188 |
+
|
| 189 |
+
def _get_optimizer(self, lr=1e-2, beta_1=0.9, beta_2=0.999, decay=1e-4):
|
| 190 |
+
return tf.keras.optimizers.Adam(lr=lr, beta_1=beta_1, beta_2=beta_2, decay=decay)
|
| 191 |
+
|
| 192 |
+
def _shuffle_data(self, filenames, labels):
|
| 193 |
+
filenames_shuffled, y_labels_shuffled = shuffle(filenames, labels)
|
| 194 |
+
return filenames_shuffled, y_labels_shuffled
|
| 195 |
+
|
| 196 |
+
def _create_generators(self):
|
| 197 |
+
fn_prefix = './file_names/' + self.dataset_name + '_'
|
| 198 |
+
# x_trains_path = fn_prefix + 'x_train_fns.npy'
|
| 199 |
+
# x_validations_path = fn_prefix + 'x_val_fns.npy'
|
| 200 |
+
|
| 201 |
+
tf_utils = TFRecordUtility(number_of_landmark=self.num_landmark)
|
| 202 |
+
|
| 203 |
+
filenames, labels = tf_utils.create_image_and_labels_name(img_path=self.img_path,
|
| 204 |
+
annotation_path=self.annotation_path)
|
| 205 |
+
filenames_shuffled, y_labels_shuffled = shuffle(filenames, labels)
|
| 206 |
+
x_train_filenames, x_val_filenames, y_train, y_val = train_test_split(
|
| 207 |
+
filenames_shuffled, y_labels_shuffled, test_size=LearningConfig.batch_size, random_state=1)
|
| 208 |
+
|
| 209 |
+
# save(x_trains_path, filenames_shuffled)
|
| 210 |
+
# save(x_validations_path, y_labels_shuffled)
|
| 211 |
+
|
| 212 |
+
# save(x_trains_path, x_train_filenames)
|
| 213 |
+
# save(x_validations_path, x_val_filenames)
|
| 214 |
+
# save(y_trains_path, y_train)
|
| 215 |
+
# save(y_validations_path, y_val)
|
| 216 |
+
|
| 217 |
+
# return filenames_shuffled, y_labels_shuffled
|
| 218 |
+
return x_train_filenames, x_val_filenames, y_train, y_val
|
| 219 |
+
|
| 220 |
+
def _create_evaluation_batch(self, x_eval_filenames, y_eval_filenames):
|
| 221 |
+
img_path = self.img_path
|
| 222 |
+
pn_tr_path = self.annotation_path
|
| 223 |
+
'''create batch data and normalize images'''
|
| 224 |
+
batch_x = x_eval_filenames[0:LearningConfig.batch_size]
|
| 225 |
+
batch_y = y_eval_filenames[0:LearningConfig.batch_size]
|
| 226 |
+
'''create img and annotations'''
|
| 227 |
+
img_batch = np.array([imread(img_path + file_name) for file_name in batch_x]) / 255.0
|
| 228 |
+
if self.dataset_name == DatasetName.cofw: # this ds is not normalized
|
| 229 |
+
pn_batch = np.array([load(pn_tr_path + file_name) for file_name in batch_y])
|
| 230 |
+
else:
|
| 231 |
+
pn_batch = np.array([self._load_and_normalize(pn_tr_path + file_name) for file_name in batch_y])
|
| 232 |
+
return img_batch, pn_batch
|
| 233 |
+
|
| 234 |
+
def _get_batch_sample(self, batch_index, x_train_filenames, y_train_filenames, model_tough_t, model_tol_t, model_student,
|
| 235 |
+
train_dif):
|
| 236 |
+
img_path = self.img_path
|
| 237 |
+
pn_tr_path = self.annotation_path
|
| 238 |
+
'''create batch data and normalize images'''
|
| 239 |
+
batch_x = x_train_filenames[
|
| 240 |
+
batch_index * LearningConfig.batch_size:(batch_index + 1) * LearningConfig.batch_size]
|
| 241 |
+
batch_y = y_train_filenames[
|
| 242 |
+
batch_index * LearningConfig.batch_size:(batch_index + 1) * LearningConfig.batch_size]
|
| 243 |
+
img_batch = np.array([imread(img_path + file_name) for file_name in batch_x]) / 255.0
|
| 244 |
+
if self.dataset_name == DatasetName.cofw: # this ds is not normalized
|
| 245 |
+
pn_batch = np.array([load(pn_tr_path + file_name) for file_name in batch_y])
|
| 246 |
+
else:
|
| 247 |
+
pn_batch = np.array([self._load_and_normalize(pn_tr_path + file_name) for file_name in batch_y])
|
| 248 |
+
'''prediction to create tough and tolerant batches'''
|
| 249 |
+
pn_batch_tough = model_tough_t.predict_on_batch(img_batch)
|
| 250 |
+
if not train_dif:
|
| 251 |
+
pn_batch_tol = model_tol_t.predict_on_batch(img_batch)
|
| 252 |
+
pn_batch_stu = None
|
| 253 |
+
else:
|
| 254 |
+
pn_batch_tol = None
|
| 255 |
+
pn_batch_stu = model_tol_t.predict_on_batch(img_batch)
|
| 256 |
+
|
| 257 |
+
|
| 258 |
+
# pn_batch_tough = 0
|
| 259 |
+
# pn_batch_tol = 0
|
| 260 |
+
|
| 261 |
+
'''test: print'''
|
| 262 |
+
# image_utility = ImageUtility()
|
| 263 |
+
# if self.dataset_name == DatasetName.cofw: # this ds is not normalized
|
| 264 |
+
# gr_s, gr_px_1, gr_Py_1 = image_utility.create_landmarks(pn_batch[0])
|
| 265 |
+
# tou_s, tou_px_1, tou_Py_1 = image_utility.create_landmarks(pn_batch_tough[0])
|
| 266 |
+
# tol_s, tol_px_1, tol_Py_1 = image_utility.create_landmarks(pn_batch_tol[0])
|
| 267 |
+
# else:
|
| 268 |
+
# gr_s, gr_px_1, gr_Py_1 = image_utility.create_landmarks_from_normalized(pn_batch[0], 224, 224, 112, 112)
|
| 269 |
+
# tou_s, tou_px_1, tou_Py_1 = image_utility.create_landmarks_from_normalized(pn_batch_tough[0], 224, 224, 112, 112)
|
| 270 |
+
# tol_s, tol_px_1, tol_Py_1 = image_utility.create_landmarks_from_normalized(pn_batch_tol[0], 224, 224, 112, 112)
|
| 271 |
+
#
|
| 272 |
+
# imgpr.print_image_arr(str(batch_index)+'pts_gt', img_batch[0], gr_px_1, gr_Py_1)
|
| 273 |
+
# imgpr.print_image_arr(str(batch_index)+'pts_t100', img_batch[0], tou_px_1, tou_Py_1)
|
| 274 |
+
# imgpr.print_image_arr(str(batch_index)+'pts_t90', img_batch[0], tol_px_1, tol_Py_1)
|
| 275 |
+
|
| 276 |
+
return img_batch, pn_batch, pn_batch_tough, pn_batch_tol, pn_batch_stu
|
| 277 |
+
|
| 278 |
+
def _load_and_normalize(self, point_path):
|
| 279 |
+
annotation = load(point_path)
|
| 280 |
+
|
| 281 |
+
"""for training we dont normalize COFW"""
|
| 282 |
+
|
| 283 |
+
'''normalize landmarks based on hyperface method'''
|
| 284 |
+
width = InputDataSize.image_input_size
|
| 285 |
+
height = InputDataSize.image_input_size
|
| 286 |
+
x_center = width / 2
|
| 287 |
+
y_center = height / 2
|
| 288 |
+
annotation_norm = []
|
| 289 |
+
for p in range(0, len(annotation), 2):
|
| 290 |
+
annotation_norm.append((x_center - annotation[p]) / width)
|
| 291 |
+
annotation_norm.append((y_center - annotation[p + 1]) / height)
|
| 292 |
+
return annotation_norm
|
teacher_trainer.py
ADDED
|
@@ -0,0 +1,215 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
from configuration import DatasetName, DatasetType, \
|
| 2 |
+
AffectnetConf, D300wConf, W300Conf, InputDataSize, LearningConfig, CofwConf, WflwConf
|
| 3 |
+
from tf_record_utility import TFRecordUtility
|
| 4 |
+
from clr_callback import CyclicLR
|
| 5 |
+
from cnn_model import CNNModel
|
| 6 |
+
from custom_Losses import Custom_losses
|
| 7 |
+
from Data_custom_generator import CustomHeatmapGenerator
|
| 8 |
+
from PW_Data_custom_generator import PWCustomHeatmapGenerator
|
| 9 |
+
from image_utility import ImageUtility
|
| 10 |
+
import img_printer as imgpr
|
| 11 |
+
|
| 12 |
+
import tensorflow as tf
|
| 13 |
+
import numpy as np
|
| 14 |
+
import matplotlib.pyplot as plt
|
| 15 |
+
import math
|
| 16 |
+
from datetime import datetime
|
| 17 |
+
from sklearn.utils import shuffle
|
| 18 |
+
from sklearn.model_selection import train_test_split
|
| 19 |
+
from numpy import save, load, asarray
|
| 20 |
+
import csv
|
| 21 |
+
from skimage.io import imread
|
| 22 |
+
import pickle
|
| 23 |
+
|
| 24 |
+
|
| 25 |
+
# tf.compat.v1.enable_eager_execution()
|
| 26 |
+
|
| 27 |
+
|
| 28 |
+
class TeacherTrainer:
|
| 29 |
+
|
| 30 |
+
def __init__(self, dataset_name, use_augmneted):
|
| 31 |
+
self.dataset_name = dataset_name
|
| 32 |
+
|
| 33 |
+
if dataset_name == DatasetName.w300:
|
| 34 |
+
self.num_landmark = D300wConf.num_of_landmarks * 2
|
| 35 |
+
if use_augmneted:
|
| 36 |
+
self.img_path = D300wConf.augmented_train_image
|
| 37 |
+
self.annotation_path = D300wConf.augmented_train_annotation
|
| 38 |
+
else:
|
| 39 |
+
self.img_path = D300wConf.no_aug_train_image
|
| 40 |
+
self.annotation_path = D300wConf.no_aug_train_annotation
|
| 41 |
+
|
| 42 |
+
if dataset_name == DatasetName.cofw:
|
| 43 |
+
self.num_landmark = CofwConf.num_of_landmarks * 2
|
| 44 |
+
self.img_path = CofwConf.augmented_train_image
|
| 45 |
+
self.annotation_path = CofwConf.augmented_train_annotation
|
| 46 |
+
|
| 47 |
+
if dataset_name == DatasetName.wflw:
|
| 48 |
+
self.num_landmark = WflwConf.num_of_landmarks * 2
|
| 49 |
+
if use_augmneted:
|
| 50 |
+
self.img_path = WflwConf.augmented_train_image
|
| 51 |
+
self.annotation_path = WflwConf.augmented_train_annotation
|
| 52 |
+
else:
|
| 53 |
+
self.img_path = WflwConf.no_aug_train_image
|
| 54 |
+
self.annotation_path = WflwConf.no_aug_train_annotation
|
| 55 |
+
|
| 56 |
+
def train(self, arch, weight_path):
|
| 57 |
+
""""""
|
| 58 |
+
'''create loss'''
|
| 59 |
+
c_loss = Custom_losses()
|
| 60 |
+
|
| 61 |
+
'''create summary writer'''
|
| 62 |
+
summary_writer = tf.summary.create_file_writer(
|
| 63 |
+
"./train_logs/fit/" + datetime.now().strftime("%Y%m%d-%H%M%S"))
|
| 64 |
+
|
| 65 |
+
'''making models'''
|
| 66 |
+
model = self.make_model(arch=arch, w_path=weight_path, is_old=False)
|
| 67 |
+
|
| 68 |
+
'''create optimizer'''
|
| 69 |
+
_lr = 1e-3
|
| 70 |
+
optimizer_student = self._get_optimizer(lr=_lr)
|
| 71 |
+
|
| 72 |
+
'''create sample generator'''
|
| 73 |
+
x_train_filenames, x_val_filenames, y_train_filenames, y_val_filenames = self._create_generators()
|
| 74 |
+
|
| 75 |
+
'''create train configuration'''
|
| 76 |
+
step_per_epoch = len(x_train_filenames) // LearningConfig.batch_size
|
| 77 |
+
|
| 78 |
+
'''start train:'''
|
| 79 |
+
for epoch in range(LearningConfig.epochs):
|
| 80 |
+
x_train_filenames, y_train_filenames = self._shuffle_data(x_train_filenames, y_train_filenames)
|
| 81 |
+
for batch_index in range(step_per_epoch):
|
| 82 |
+
'''load annotation and images'''
|
| 83 |
+
images, annotation_gr = self._get_batch_sample(
|
| 84 |
+
batch_index=batch_index, x_train_filenames=x_train_filenames,
|
| 85 |
+
y_train_filenames=y_train_filenames, model=model)
|
| 86 |
+
'''convert to tensor'''
|
| 87 |
+
images = tf.cast(images, tf.float32)
|
| 88 |
+
annotation_gr = tf.cast(annotation_gr, tf.float32)
|
| 89 |
+
|
| 90 |
+
'''train step'''
|
| 91 |
+
self.train_step(epoch=epoch, step=batch_index, total_steps=step_per_epoch, images=images,
|
| 92 |
+
model=model,
|
| 93 |
+
annotation_gr=annotation_gr,
|
| 94 |
+
optimizer=optimizer_student,
|
| 95 |
+
summary_writer=summary_writer, c_loss=c_loss)
|
| 96 |
+
'''evaluating part'''
|
| 97 |
+
img_batch_eval, pn_batch_eval = self._create_evaluation_batch(x_val_filenames, y_val_filenames)
|
| 98 |
+
# loss_eval, loss_eval_tol_dif_stu, loss_eval_tol_dif_gt, loss_eval_tou_dif_stu, loss_eval_tou_dif_gt = \
|
| 99 |
+
loss_eval = self._eval_model(img_batch_eval, pn_batch_eval, model)
|
| 100 |
+
with summary_writer.as_default():
|
| 101 |
+
tf.summary.scalar('Eval-LOSS', loss_eval, step=epoch)
|
| 102 |
+
'''save weights'''
|
| 103 |
+
model.save(
|
| 104 |
+
'./models/teacher_model_' + str(epoch) + '_' + self.dataset_name + '_' + str(loss_eval) + '.h5')
|
| 105 |
+
|
| 106 |
+
# @tf.function
|
| 107 |
+
def train_step(self, epoch, step, total_steps, images,
|
| 108 |
+
model, annotation_gr,
|
| 109 |
+
optimizer, summary_writer, c_loss):
|
| 110 |
+
with tf.GradientTape() as tape:
|
| 111 |
+
'''create annotation_predicted'''
|
| 112 |
+
annotation_predicted = model(images, training=True)
|
| 113 |
+
'''calculate loss'''
|
| 114 |
+
loss = c_loss.MSE(x_pr=annotation_predicted, x_gt=annotation_gr)
|
| 115 |
+
'''calculate gradient'''
|
| 116 |
+
gradients = tape.gradient(loss, model.trainable_variables)
|
| 117 |
+
'''apply Gradients:'''
|
| 118 |
+
optimizer.apply_gradients(zip(gradients, model.trainable_variables))
|
| 119 |
+
'''printing loss Values: '''
|
| 120 |
+
tf.print("->EPOCH: ", str(epoch), "->STEP: ", str(step) + '/' + str(total_steps), ' -> : LOSS: ', loss)
|
| 121 |
+
with summary_writer.as_default():
|
| 122 |
+
tf.summary.scalar('LOSS', loss, step=epoch)
|
| 123 |
+
|
| 124 |
+
def make_model(self, arch, w_path, is_old=False):
|
| 125 |
+
cnn = CNNModel()
|
| 126 |
+
model = cnn.get_model(arch=arch, output_len=self.num_landmark, input_tensor=None, weight_path=w_path,
|
| 127 |
+
is_old=is_old)
|
| 128 |
+
if w_path is not None and arch != 'mobileNetV2_d' and not is_old:
|
| 129 |
+
model.load_weights(w_path)
|
| 130 |
+
return model
|
| 131 |
+
|
| 132 |
+
def _eval_model(self, img_batch_eval, pn_batch_eval, model):
|
| 133 |
+
# annotation_predicted, pr_tol_dif_stu, pr_tol_dif_gt, pr_tou_dif_stu, pr_tou_dif_gt = model(img_batch_eval)
|
| 134 |
+
annotation_predicted = model(img_batch_eval)
|
| 135 |
+
loss_eval = np.array(tf.reduce_mean(tf.abs(pn_batch_eval - annotation_predicted)))
|
| 136 |
+
# loss_eval_tol_dif_stu = np.array(tf.reduce_mean(tf.abs(pn_batch_eval - annotation_predicted)))
|
| 137 |
+
# loss_eval_tol_dif_gt = np.array(tf.reduce_mean(tf.abs(pn_batch_eval - annotation_predicted)))
|
| 138 |
+
# loss_eval_tou_dif_stu = np.array(tf.reduce_mean(tf.abs(pn_batch_eval - annotation_predicted)))
|
| 139 |
+
# loss_eval_tou_dif_gt = np.array(tf.reduce_mean(tf.abs(pn_batch_eval - annotation_predicted)))
|
| 140 |
+
# return loss_eval, loss_eval_tol_dif_stu, loss_eval_tol_dif_gt, loss_eval_tou_dif_stu, loss_eval_tou_dif_gt
|
| 141 |
+
return loss_eval
|
| 142 |
+
|
| 143 |
+
def _get_optimizer(self, lr=1e-2, beta_1=0.9, beta_2=0.999, decay=1e-4):
|
| 144 |
+
return tf.keras.optimizers.Adam(lr=lr, beta_1=beta_1, beta_2=beta_2, decay=decay)
|
| 145 |
+
|
| 146 |
+
def _shuffle_data(self, filenames, labels):
|
| 147 |
+
filenames_shuffled, y_labels_shuffled = shuffle(filenames, labels)
|
| 148 |
+
return filenames_shuffled, y_labels_shuffled
|
| 149 |
+
|
| 150 |
+
def _create_generators(self):
|
| 151 |
+
fn_prefix = './file_names/' + self.dataset_name + '_'
|
| 152 |
+
# x_trains_path = fn_prefix + 'x_train_fns.npy'
|
| 153 |
+
# x_validations_path = fn_prefix + 'x_val_fns.npy'
|
| 154 |
+
|
| 155 |
+
tf_utils = TFRecordUtility(number_of_landmark=self.num_landmark)
|
| 156 |
+
|
| 157 |
+
filenames, labels = tf_utils.create_image_and_labels_name(img_path=self.img_path,
|
| 158 |
+
annotation_path=self.annotation_path)
|
| 159 |
+
filenames_shuffled, y_labels_shuffled = shuffle(filenames, labels)
|
| 160 |
+
x_train_filenames, x_val_filenames, y_train, y_val = train_test_split(
|
| 161 |
+
filenames_shuffled, y_labels_shuffled, test_size=LearningConfig.batch_size, random_state=1)
|
| 162 |
+
|
| 163 |
+
# save(x_trains_path, filenames_shuffled)
|
| 164 |
+
# save(x_validations_path, y_labels_shuffled)
|
| 165 |
+
|
| 166 |
+
# save(x_trains_path, x_train_filenames)
|
| 167 |
+
# save(x_validations_path, x_val_filenames)
|
| 168 |
+
# save(y_trains_path, y_train)
|
| 169 |
+
# save(y_validations_path, y_val)
|
| 170 |
+
|
| 171 |
+
# return filenames_shuffled, y_labels_shuffled
|
| 172 |
+
return x_train_filenames, x_val_filenames, y_train, y_val
|
| 173 |
+
|
| 174 |
+
def _create_evaluation_batch(self, x_eval_filenames, y_eval_filenames):
|
| 175 |
+
img_path = self.img_path
|
| 176 |
+
pn_tr_path = self.annotation_path
|
| 177 |
+
'''create batch data and normalize images'''
|
| 178 |
+
batch_x = x_eval_filenames[0:LearningConfig.batch_size]
|
| 179 |
+
batch_y = y_eval_filenames[0:LearningConfig.batch_size]
|
| 180 |
+
'''create img and annotations'''
|
| 181 |
+
img_batch = np.array([imread(img_path + file_name) for file_name in batch_x]) / 255.0
|
| 182 |
+
if self.dataset_name == DatasetName.cofw: # this ds is not normalized
|
| 183 |
+
pn_batch = np.array([load(pn_tr_path + file_name) for file_name in batch_y])
|
| 184 |
+
else:
|
| 185 |
+
pn_batch = np.array([self._load_and_normalize(pn_tr_path + file_name) for file_name in batch_y])
|
| 186 |
+
return img_batch, pn_batch
|
| 187 |
+
|
| 188 |
+
def _get_batch_sample(self, batch_index, x_train_filenames, y_train_filenames):
|
| 189 |
+
img_path = self.img_path
|
| 190 |
+
pn_tr_path = self.annotation_path
|
| 191 |
+
'''create batch data and normalize images'''
|
| 192 |
+
batch_x = x_train_filenames[
|
| 193 |
+
batch_index * LearningConfig.batch_size:(batch_index + 1) * LearningConfig.batch_size]
|
| 194 |
+
batch_y = y_train_filenames[
|
| 195 |
+
batch_index * LearningConfig.batch_size:(batch_index + 1) * LearningConfig.batch_size]
|
| 196 |
+
img_batch = np.array([imread(img_path + file_name) for file_name in batch_x]) / 255.0
|
| 197 |
+
pn_batch = np.array([self._load_and_normalize(pn_tr_path + file_name) for file_name in batch_y])
|
| 198 |
+
|
| 199 |
+
return img_batch, pn_batch
|
| 200 |
+
|
| 201 |
+
def _load_and_normalize(self, point_path):
|
| 202 |
+
annotation = load(point_path)
|
| 203 |
+
|
| 204 |
+
"""for training we dont normalize COFW"""
|
| 205 |
+
|
| 206 |
+
'''normalize landmarks based on hyperface method'''
|
| 207 |
+
width = InputDataSize.image_input_size
|
| 208 |
+
height = InputDataSize.image_input_size
|
| 209 |
+
x_center = width / 2
|
| 210 |
+
y_center = height / 2
|
| 211 |
+
annotation_norm = []
|
| 212 |
+
for p in range(0, len(annotation), 2):
|
| 213 |
+
annotation_norm.append((x_center - annotation[p]) / width)
|
| 214 |
+
annotation_norm.append((y_center - annotation[p + 1]) / height)
|
| 215 |
+
return annotation_norm
|
test.py
ADDED
|
@@ -0,0 +1,40 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
from configuration import DatasetName, DatasetType, \
|
| 2 |
+
AffectnetConf, D300wConf, W300Conf, InputDataSize, LearningConfig, CofwConf, WflwConf
|
| 3 |
+
from tf_record_utility import TFRecordUtility
|
| 4 |
+
from image_utility import ImageUtility
|
| 5 |
+
from skimage.transform import resize
|
| 6 |
+
import numpy as np
|
| 7 |
+
import math
|
| 8 |
+
|
| 9 |
+
import cv2
|
| 10 |
+
import os.path
|
| 11 |
+
import scipy.io as sio
|
| 12 |
+
from cnn_model import CNNModel
|
| 13 |
+
import img_printer as imgpr
|
| 14 |
+
from tqdm import tqdm
|
| 15 |
+
|
| 16 |
+
|
| 17 |
+
class Test:
|
| 18 |
+
def __init__(self, dataset_name, arch, num_output_layers, weight_fname, has_pose=False):
|
| 19 |
+
self.dataset_name = dataset_name
|
| 20 |
+
self.has_pose = has_pose
|
| 21 |
+
|
| 22 |
+
if dataset_name == DatasetName.w300:
|
| 23 |
+
self.output_len = D300wConf.num_of_landmarks * 2
|
| 24 |
+
elif dataset_name == DatasetName.cofw:
|
| 25 |
+
self.output_len = CofwConf.num_of_landmarks * 2
|
| 26 |
+
elif dataset_name == DatasetName.wflw:
|
| 27 |
+
self.output_len = WflwConf.num_of_landmarks * 2
|
| 28 |
+
|
| 29 |
+
cnn = CNNModel()
|
| 30 |
+
model = cnn.get_model(arch=arch, input_tensor=None, output_len=self.output_len)
|
| 31 |
+
|
| 32 |
+
model.load_weights(weight_fname)
|
| 33 |
+
|
| 34 |
+
img = None # load a cropped image
|
| 35 |
+
|
| 36 |
+
image_utility = ImageUtility()
|
| 37 |
+
pose_predicted = []
|
| 38 |
+
image = np.expand_dims(img, axis=0)
|
| 39 |
+
|
| 40 |
+
pose_predicted = model.predict(image)[1][0]
|